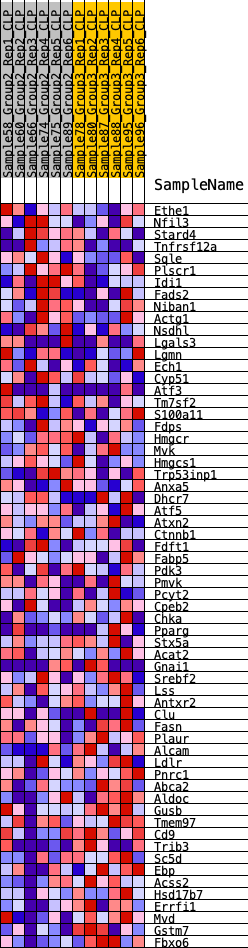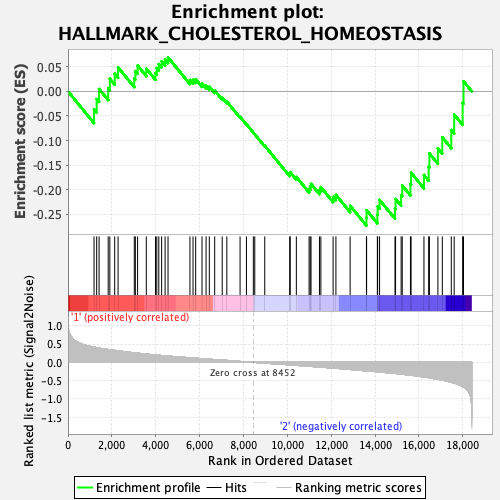
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group2_versus_Group3.Basophil_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | Basophil_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_CHOLESTEROL_HOMEOSTASIS |
| Enrichment Score (ES) | -0.27301338 |
| Normalized Enrichment Score (NES) | -0.9505899 |
| Nominal p-value | 0.537797 |
| FDR q-value | 0.8676764 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ethe1 | 1187 | 0.414 | -0.0364 | No |
| 2 | Nfil3 | 1307 | 0.400 | -0.0156 | No |
| 3 | Stard4 | 1417 | 0.388 | 0.0049 | No |
| 4 | Tnfrsf12a | 1832 | 0.350 | 0.0062 | No |
| 5 | Sqle | 1907 | 0.344 | 0.0256 | No |
| 6 | Plscr1 | 2129 | 0.325 | 0.0358 | No |
| 7 | Idi1 | 2282 | 0.315 | 0.0489 | No |
| 8 | Fads2 | 3021 | 0.258 | 0.0263 | No |
| 9 | Niban1 | 3070 | 0.256 | 0.0411 | No |
| 10 | Actg1 | 3173 | 0.248 | 0.0525 | No |
| 11 | Nsdhl | 3569 | 0.224 | 0.0463 | No |
| 12 | Lgals3 | 3988 | 0.200 | 0.0371 | No |
| 13 | Lgmn | 4047 | 0.197 | 0.0474 | No |
| 14 | Ech1 | 4139 | 0.192 | 0.0555 | No |
| 15 | Cyp51 | 4268 | 0.185 | 0.0612 | No |
| 16 | Atf3 | 4426 | 0.177 | 0.0647 | No |
| 17 | Tm7sf2 | 4560 | 0.171 | 0.0691 | No |
| 18 | S100a11 | 5559 | 0.125 | 0.0232 | No |
| 19 | Fdps | 5700 | 0.117 | 0.0236 | No |
| 20 | Hmgcr | 5820 | 0.112 | 0.0247 | No |
| 21 | Mvk | 6109 | 0.099 | 0.0158 | No |
| 22 | Hmgcs1 | 6294 | 0.091 | 0.0120 | No |
| 23 | Trp53inp1 | 6442 | 0.085 | 0.0098 | No |
| 24 | Anxa5 | 6687 | 0.075 | 0.0016 | No |
| 25 | Dhcr7 | 7030 | 0.061 | -0.0129 | No |
| 26 | Atf5 | 7240 | 0.052 | -0.0207 | No |
| 27 | Atxn2 | 7844 | 0.024 | -0.0519 | No |
| 28 | Ctnnb1 | 8135 | 0.012 | -0.0669 | No |
| 29 | Fdft1 | 8449 | 0.000 | -0.0839 | No |
| 30 | Fabp5 | 8514 | -0.002 | -0.0873 | No |
| 31 | Pdk3 | 8969 | -0.020 | -0.1106 | No |
| 32 | Pmvk | 10106 | -0.068 | -0.1679 | No |
| 33 | Pcyt2 | 10129 | -0.069 | -0.1644 | No |
| 34 | Cpeb2 | 10409 | -0.082 | -0.1741 | No |
| 35 | Chka | 10990 | -0.107 | -0.1984 | No |
| 36 | Pparg | 11039 | -0.109 | -0.1936 | No |
| 37 | Stx5a | 11078 | -0.110 | -0.1881 | No |
| 38 | Acat2 | 11463 | -0.128 | -0.2003 | No |
| 39 | Gnai1 | 11527 | -0.131 | -0.1948 | No |
| 40 | Srebf2 | 12085 | -0.155 | -0.2146 | No |
| 41 | Lss | 12213 | -0.162 | -0.2105 | No |
| 42 | Antxr2 | 12861 | -0.194 | -0.2325 | No |
| 43 | Clu | 13605 | -0.232 | -0.2572 | Yes |
| 44 | Fasn | 13612 | -0.233 | -0.2416 | Yes |
| 45 | Plaur | 14102 | -0.257 | -0.2508 | Yes |
| 46 | Alcam | 14115 | -0.257 | -0.2339 | Yes |
| 47 | Ldlr | 14203 | -0.262 | -0.2207 | Yes |
| 48 | Pnrc1 | 14907 | -0.303 | -0.2384 | Yes |
| 49 | Abca2 | 14932 | -0.305 | -0.2189 | Yes |
| 50 | Aldoc | 15186 | -0.321 | -0.2108 | Yes |
| 51 | Gusb | 15237 | -0.324 | -0.1914 | Yes |
| 52 | Tmem97 | 15611 | -0.350 | -0.1879 | Yes |
| 53 | Cd9 | 15640 | -0.351 | -0.1654 | Yes |
| 54 | Trib3 | 16226 | -0.399 | -0.1701 | Yes |
| 55 | Sc5d | 16441 | -0.419 | -0.1532 | Yes |
| 56 | Ebp | 16475 | -0.422 | -0.1262 | Yes |
| 57 | Acss2 | 16862 | -0.464 | -0.1156 | Yes |
| 58 | Hsd17b7 | 17063 | -0.484 | -0.0935 | Yes |
| 59 | Errfi1 | 17472 | -0.545 | -0.0786 | Yes |
| 60 | Mvd | 17604 | -0.566 | -0.0471 | Yes |
| 61 | Gstm7 | 17990 | -0.658 | -0.0232 | Yes |
| 62 | Fbxo6 | 18029 | -0.673 | 0.0207 | Yes |