Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group2_versus_Group3.Basophil_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | Basophil_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
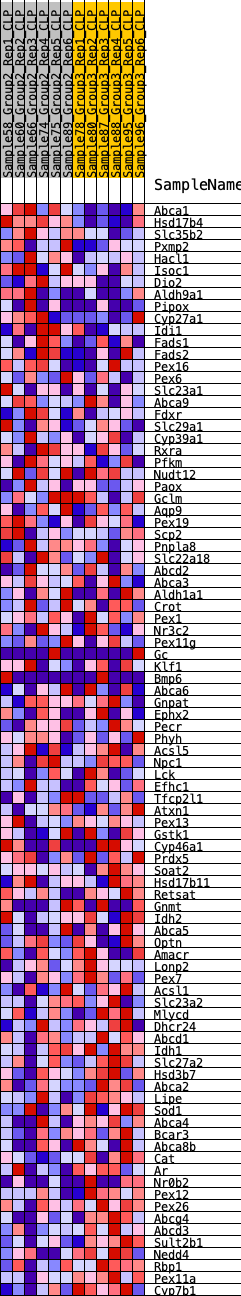
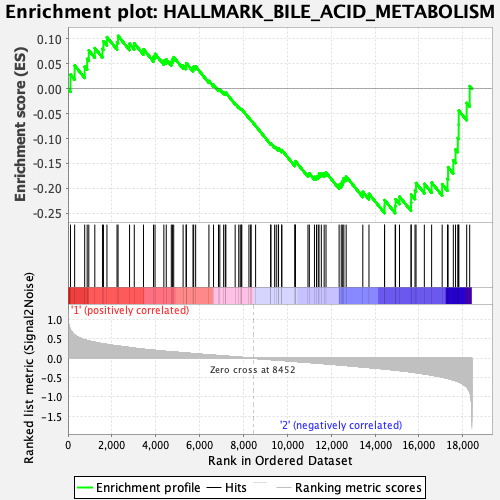
| GeneSet | HALLMARK_BILE_ACID_METABOLISM |
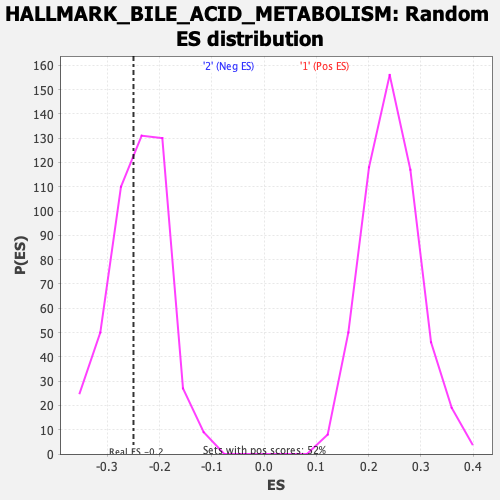
| Enrichment Score (ES) | -0.24975348 |
| Normalized Enrichment Score (NES) | -1.04358 |
| Nominal p-value | 0.39834026 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.984 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Abca1 | 113 | 0.733 | 0.0286 | No |
| 2 | Hsd17b4 | 303 | 0.598 | 0.0467 | No |
| 3 | Slc35b2 | 761 | 0.476 | 0.0443 | No |
| 4 | Pxmp2 | 874 | 0.458 | 0.0599 | No |
| 5 | Hacl1 | 948 | 0.447 | 0.0771 | No |
| 6 | Isoc1 | 1222 | 0.411 | 0.0817 | No |
| 7 | Dio2 | 1571 | 0.373 | 0.0804 | No |
| 8 | Aldh9a1 | 1616 | 0.369 | 0.0955 | No |
| 9 | Pipox | 1775 | 0.356 | 0.1037 | No |
| 10 | Cyp27a1 | 2235 | 0.318 | 0.0938 | No |
| 11 | Idi1 | 2282 | 0.315 | 0.1062 | No |
| 12 | Fads1 | 2806 | 0.275 | 0.0906 | No |
| 13 | Fads2 | 3021 | 0.258 | 0.0912 | No |
| 14 | Pex16 | 3441 | 0.232 | 0.0794 | No |
| 15 | Pex6 | 3899 | 0.206 | 0.0642 | No |
| 16 | Slc23a1 | 3968 | 0.201 | 0.0700 | No |
| 17 | Abca9 | 4370 | 0.179 | 0.0566 | No |
| 18 | Fdxr | 4479 | 0.174 | 0.0590 | No |
| 19 | Slc29a1 | 4709 | 0.163 | 0.0542 | No |
| 20 | Cyp39a1 | 4761 | 0.161 | 0.0591 | No |
| 21 | Rxra | 4819 | 0.159 | 0.0635 | No |
| 22 | Pfkm | 5247 | 0.139 | 0.0468 | No |
| 23 | Nudt12 | 5382 | 0.132 | 0.0457 | No |
| 24 | Paox | 5396 | 0.132 | 0.0513 | No |
| 25 | Gclm | 5698 | 0.117 | 0.0404 | No |
| 26 | Aqp9 | 5729 | 0.116 | 0.0443 | No |
| 27 | Pex19 | 5819 | 0.112 | 0.0447 | No |
| 28 | Scp2 | 6424 | 0.086 | 0.0158 | No |
| 29 | Pnpla8 | 6634 | 0.076 | 0.0080 | No |
| 30 | Slc22a18 | 6860 | 0.068 | -0.0010 | No |
| 31 | Abcd2 | 6921 | 0.065 | -0.0012 | No |
| 32 | Abca3 | 7095 | 0.058 | -0.0079 | No |
| 33 | Aldh1a1 | 7170 | 0.055 | -0.0093 | No |
| 34 | Crot | 7196 | 0.054 | -0.0081 | No |
| 35 | Pex1 | 7620 | 0.034 | -0.0296 | No |
| 36 | Nr3c2 | 7784 | 0.027 | -0.0372 | No |
| 37 | Pex11g | 7851 | 0.024 | -0.0397 | No |
| 38 | Gc | 7899 | 0.022 | -0.0412 | No |
| 39 | Klf1 | 7923 | 0.022 | -0.0414 | No |
| 40 | Bmp6 | 8248 | 0.008 | -0.0587 | No |
| 41 | Abca6 | 8327 | 0.005 | -0.0627 | No |
| 42 | Gnpat | 8347 | 0.004 | -0.0635 | No |
| 43 | Ephx2 | 8552 | -0.003 | -0.0745 | No |
| 44 | Pecr | 9239 | -0.032 | -0.1104 | No |
| 45 | Phyh | 9250 | -0.032 | -0.1094 | No |
| 46 | Acsl5 | 9420 | -0.040 | -0.1168 | No |
| 47 | Npc1 | 9494 | -0.043 | -0.1187 | No |
| 48 | Lck | 9586 | -0.046 | -0.1215 | No |
| 49 | Efhc1 | 9598 | -0.046 | -0.1199 | No |
| 50 | Tfcp2l1 | 9738 | -0.052 | -0.1251 | No |
| 51 | Atxn1 | 9749 | -0.053 | -0.1231 | No |
| 52 | Pex13 | 10342 | -0.079 | -0.1517 | No |
| 53 | Gstk1 | 10352 | -0.079 | -0.1484 | No |
| 54 | Cyp46a1 | 10367 | -0.080 | -0.1454 | No |
| 55 | Prdx5 | 10939 | -0.105 | -0.1716 | No |
| 56 | Soat2 | 11002 | -0.107 | -0.1699 | No |
| 57 | Hsd17b11 | 11235 | -0.117 | -0.1770 | No |
| 58 | Retsat | 11328 | -0.121 | -0.1763 | No |
| 59 | Gnmt | 11406 | -0.125 | -0.1745 | No |
| 60 | Idh2 | 11442 | -0.127 | -0.1705 | No |
| 61 | Abca5 | 11548 | -0.132 | -0.1699 | No |
| 62 | Optn | 11677 | -0.137 | -0.1704 | No |
| 63 | Amacr | 11761 | -0.141 | -0.1683 | No |
| 64 | Lonp2 | 12359 | -0.169 | -0.1928 | No |
| 65 | Pex7 | 12456 | -0.174 | -0.1898 | No |
| 66 | Acsl1 | 12522 | -0.177 | -0.1850 | No |
| 67 | Slc23a2 | 12575 | -0.180 | -0.1793 | No |
| 68 | Mlycd | 12680 | -0.185 | -0.1762 | No |
| 69 | Dhcr24 | 13439 | -0.224 | -0.2069 | No |
| 70 | Abcd1 | 13720 | -0.238 | -0.2109 | No |
| 71 | Idh1 | 14430 | -0.276 | -0.2365 | Yes |
| 72 | Slc27a2 | 14432 | -0.276 | -0.2235 | Yes |
| 73 | Hsd3b7 | 14914 | -0.304 | -0.2353 | Yes |
| 74 | Abca2 | 14932 | -0.305 | -0.2218 | Yes |
| 75 | Lipe | 15111 | -0.316 | -0.2165 | Yes |
| 76 | Sod1 | 15637 | -0.351 | -0.2285 | Yes |
| 77 | Abca4 | 15645 | -0.351 | -0.2123 | Yes |
| 78 | Bcar3 | 15819 | -0.366 | -0.2043 | Yes |
| 79 | Abca8b | 15865 | -0.370 | -0.1892 | Yes |
| 80 | Cat | 16244 | -0.401 | -0.1909 | Yes |
| 81 | Ar | 16576 | -0.434 | -0.1884 | Yes |
| 82 | Nr0b2 | 17057 | -0.484 | -0.1916 | Yes |
| 83 | Pex12 | 17299 | -0.514 | -0.1804 | Yes |
| 84 | Pex26 | 17327 | -0.518 | -0.1574 | Yes |
| 85 | Abcg4 | 17565 | -0.560 | -0.1437 | Yes |
| 86 | Abcd3 | 17666 | -0.576 | -0.1219 | Yes |
| 87 | Sult2b1 | 17769 | -0.599 | -0.0990 | Yes |
| 88 | Nedd4 | 17809 | -0.606 | -0.0724 | Yes |
| 89 | Rbp1 | 17812 | -0.606 | -0.0438 | Yes |
| 90 | Pex11a | 18175 | -0.740 | -0.0284 | Yes |
| 91 | Cyp7b1 | 18311 | -0.867 | 0.0053 | Yes |