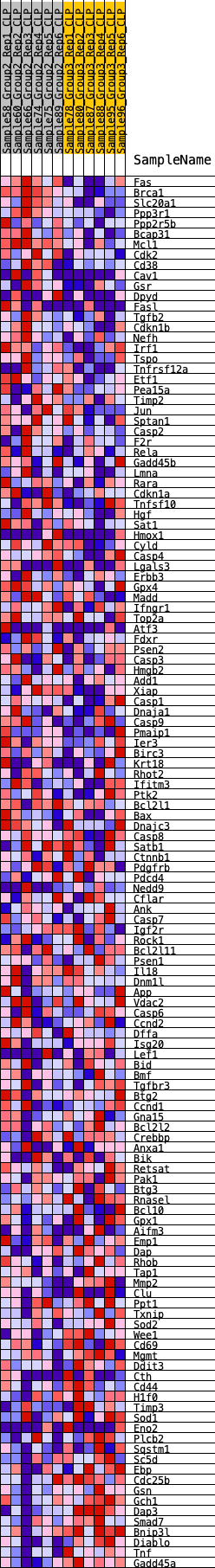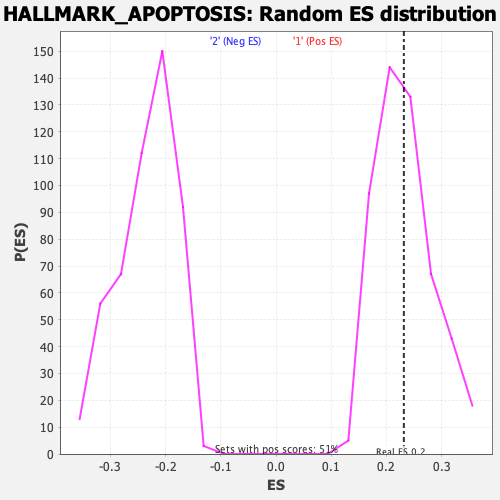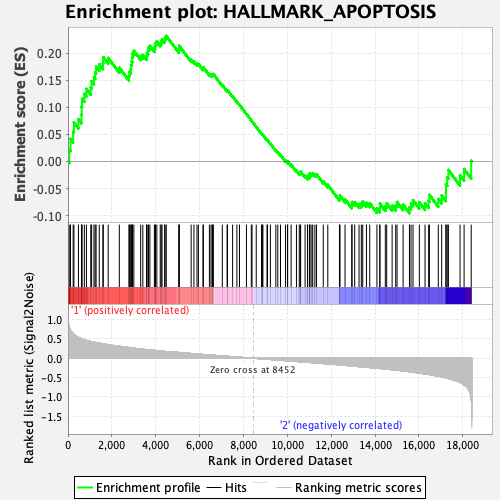
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group2_versus_Group3.Basophil_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | Basophil_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_APOPTOSIS |
| Enrichment Score (ES) | 0.2317242 |
| Normalized Enrichment Score (NES) | 1.0002114 |
| Nominal p-value | 0.43786982 |
| FDR q-value | 0.8755111 |
| FWER p-Value | 0.999 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Fas | 55 | 0.810 | 0.0222 | Yes |
| 2 | Brca1 | 114 | 0.730 | 0.0417 | Yes |
| 3 | Slc20a1 | 235 | 0.638 | 0.0549 | Yes |
| 4 | Ppp3r1 | 270 | 0.614 | 0.0721 | Yes |
| 5 | Ppp2r5b | 476 | 0.542 | 0.0778 | Yes |
| 6 | Bcap31 | 614 | 0.503 | 0.0859 | Yes |
| 7 | Mcl1 | 622 | 0.501 | 0.1011 | Yes |
| 8 | Cdk2 | 642 | 0.497 | 0.1155 | Yes |
| 9 | Cd38 | 745 | 0.477 | 0.1248 | Yes |
| 10 | Cav1 | 840 | 0.464 | 0.1340 | Yes |
| 11 | Gsr | 1039 | 0.434 | 0.1367 | Yes |
| 12 | Dpyd | 1076 | 0.429 | 0.1480 | Yes |
| 13 | Fasl | 1183 | 0.415 | 0.1551 | Yes |
| 14 | Tgfb2 | 1238 | 0.408 | 0.1649 | Yes |
| 15 | Cdkn1b | 1281 | 0.403 | 0.1751 | Yes |
| 16 | Nefh | 1425 | 0.387 | 0.1793 | Yes |
| 17 | Irf1 | 1593 | 0.371 | 0.1817 | Yes |
| 18 | Tspo | 1606 | 0.370 | 0.1925 | Yes |
| 19 | Tnfrsf12a | 1832 | 0.350 | 0.1911 | Yes |
| 20 | Etf1 | 2339 | 0.311 | 0.1731 | Yes |
| 21 | Pea15a | 2778 | 0.277 | 0.1577 | Yes |
| 22 | Timp2 | 2799 | 0.275 | 0.1652 | Yes |
| 23 | Jun | 2863 | 0.271 | 0.1702 | Yes |
| 24 | Sptan1 | 2871 | 0.271 | 0.1782 | Yes |
| 25 | Casp2 | 2904 | 0.268 | 0.1848 | Yes |
| 26 | F2r | 2930 | 0.265 | 0.1916 | Yes |
| 27 | Rela | 2938 | 0.264 | 0.1995 | Yes |
| 28 | Gadd45b | 3001 | 0.260 | 0.2041 | Yes |
| 29 | Lmna | 3313 | 0.240 | 0.1946 | Yes |
| 30 | Rara | 3411 | 0.234 | 0.1965 | Yes |
| 31 | Cdkn1a | 3579 | 0.224 | 0.1944 | Yes |
| 32 | Tnfsf10 | 3599 | 0.223 | 0.2002 | Yes |
| 33 | Hgf | 3652 | 0.219 | 0.2042 | Yes |
| 34 | Sat1 | 3669 | 0.218 | 0.2101 | Yes |
| 35 | Hmox1 | 3733 | 0.215 | 0.2133 | Yes |
| 36 | Cyld | 3938 | 0.203 | 0.2085 | Yes |
| 37 | Casp4 | 3969 | 0.201 | 0.2131 | Yes |
| 38 | Lgals3 | 3988 | 0.200 | 0.2183 | Yes |
| 39 | Erbb3 | 4049 | 0.197 | 0.2212 | Yes |
| 40 | Gpx4 | 4204 | 0.188 | 0.2186 | Yes |
| 41 | Madd | 4249 | 0.186 | 0.2220 | Yes |
| 42 | Ifngr1 | 4286 | 0.184 | 0.2257 | Yes |
| 43 | Top2a | 4405 | 0.178 | 0.2248 | Yes |
| 44 | Atf3 | 4426 | 0.177 | 0.2292 | Yes |
| 45 | Fdxr | 4479 | 0.174 | 0.2317 | Yes |
| 46 | Psen2 | 5052 | 0.149 | 0.2051 | No |
| 47 | Casp3 | 5067 | 0.148 | 0.2089 | No |
| 48 | Hmgb2 | 5068 | 0.148 | 0.2135 | No |
| 49 | Add1 | 5614 | 0.122 | 0.1875 | No |
| 50 | Xiap | 5741 | 0.116 | 0.1842 | No |
| 51 | Casp1 | 5881 | 0.109 | 0.1800 | No |
| 52 | Dnaja1 | 5944 | 0.107 | 0.1799 | No |
| 53 | Casp9 | 6153 | 0.097 | 0.1715 | No |
| 54 | Pmaip1 | 6170 | 0.096 | 0.1736 | No |
| 55 | Ier3 | 6445 | 0.085 | 0.1613 | No |
| 56 | Birc3 | 6518 | 0.081 | 0.1599 | No |
| 57 | Krt18 | 6573 | 0.079 | 0.1594 | No |
| 58 | Rhot2 | 6587 | 0.079 | 0.1611 | No |
| 59 | Ifitm3 | 6628 | 0.077 | 0.1613 | No |
| 60 | Ptk2 | 7035 | 0.061 | 0.1410 | No |
| 61 | Bcl2l1 | 7253 | 0.052 | 0.1307 | No |
| 62 | Bax | 7270 | 0.051 | 0.1314 | No |
| 63 | Dnajc3 | 7504 | 0.040 | 0.1199 | No |
| 64 | Casp8 | 7699 | 0.031 | 0.1103 | No |
| 65 | Satb1 | 7817 | 0.025 | 0.1046 | No |
| 66 | Ctnnb1 | 8135 | 0.012 | 0.0877 | No |
| 67 | Pdgfrb | 8356 | 0.004 | 0.0758 | No |
| 68 | Pdcd4 | 8389 | 0.002 | 0.0741 | No |
| 69 | Nedd9 | 8581 | -0.005 | 0.0638 | No |
| 70 | Cflar | 8820 | -0.015 | 0.0512 | No |
| 71 | Ank | 8875 | -0.017 | 0.0488 | No |
| 72 | Casp7 | 8879 | -0.017 | 0.0491 | No |
| 73 | Igf2r | 9072 | -0.025 | 0.0394 | No |
| 74 | Rock1 | 9094 | -0.026 | 0.0391 | No |
| 75 | Bcl2l11 | 9226 | -0.032 | 0.0329 | No |
| 76 | Psen1 | 9474 | -0.042 | 0.0207 | No |
| 77 | Il18 | 9564 | -0.045 | 0.0172 | No |
| 78 | Dnm1l | 9687 | -0.050 | 0.0121 | No |
| 79 | App | 9913 | -0.060 | 0.0016 | No |
| 80 | Vdac2 | 10004 | -0.064 | -0.0013 | No |
| 81 | Casp6 | 10014 | -0.064 | 0.0002 | No |
| 82 | Ccnd2 | 10171 | -0.071 | -0.0062 | No |
| 83 | Dffa | 10414 | -0.082 | -0.0168 | No |
| 84 | Isg20 | 10541 | -0.088 | -0.0210 | No |
| 85 | Lef1 | 10590 | -0.090 | -0.0209 | No |
| 86 | Bid | 10605 | -0.090 | -0.0188 | No |
| 87 | Bmf | 10809 | -0.099 | -0.0269 | No |
| 88 | Tgfbr3 | 10920 | -0.104 | -0.0296 | No |
| 89 | Btg2 | 10922 | -0.104 | -0.0264 | No |
| 90 | Ccnd1 | 11013 | -0.108 | -0.0280 | No |
| 91 | Gna15 | 11022 | -0.108 | -0.0251 | No |
| 92 | Bcl2l2 | 11025 | -0.108 | -0.0218 | No |
| 93 | Crebbp | 11118 | -0.112 | -0.0234 | No |
| 94 | Anxa1 | 11158 | -0.114 | -0.0220 | No |
| 95 | Bik | 11255 | -0.118 | -0.0236 | No |
| 96 | Retsat | 11328 | -0.121 | -0.0237 | No |
| 97 | Pak1 | 11636 | -0.135 | -0.0363 | No |
| 98 | Btg3 | 11843 | -0.145 | -0.0431 | No |
| 99 | Rnasel | 12374 | -0.170 | -0.0668 | No |
| 100 | Bcl10 | 12400 | -0.171 | -0.0629 | No |
| 101 | Gpx1 | 12629 | -0.183 | -0.0697 | No |
| 102 | Aifm3 | 12935 | -0.198 | -0.0802 | No |
| 103 | Emp1 | 12949 | -0.199 | -0.0748 | No |
| 104 | Dap | 13069 | -0.206 | -0.0748 | No |
| 105 | Rhob | 13267 | -0.215 | -0.0789 | No |
| 106 | Tap1 | 13371 | -0.220 | -0.0777 | No |
| 107 | Mmp2 | 13435 | -0.224 | -0.0742 | No |
| 108 | Clu | 13605 | -0.232 | -0.0762 | No |
| 109 | Ppt1 | 13755 | -0.240 | -0.0769 | No |
| 110 | Txnip | 14082 | -0.256 | -0.0868 | No |
| 111 | Sod2 | 14215 | -0.263 | -0.0859 | No |
| 112 | Wee1 | 14218 | -0.263 | -0.0778 | No |
| 113 | Cd69 | 14465 | -0.278 | -0.0826 | No |
| 114 | Mgmt | 14528 | -0.281 | -0.0773 | No |
| 115 | Ddit3 | 14775 | -0.295 | -0.0816 | No |
| 116 | Cth | 14939 | -0.306 | -0.0810 | No |
| 117 | Cd44 | 15000 | -0.309 | -0.0747 | No |
| 118 | H1f0 | 15276 | -0.326 | -0.0796 | No |
| 119 | Timp3 | 15565 | -0.346 | -0.0846 | No |
| 120 | Sod1 | 15637 | -0.351 | -0.0776 | No |
| 121 | Eno2 | 15728 | -0.359 | -0.0713 | No |
| 122 | Plcb2 | 16016 | -0.381 | -0.0752 | No |
| 123 | Sqstm1 | 16276 | -0.404 | -0.0768 | No |
| 124 | Sc5d | 16441 | -0.419 | -0.0728 | No |
| 125 | Ebp | 16475 | -0.422 | -0.0615 | No |
| 126 | Cdc25b | 16880 | -0.465 | -0.0691 | No |
| 127 | Gsn | 17034 | -0.481 | -0.0626 | No |
| 128 | Gch1 | 17228 | -0.504 | -0.0575 | No |
| 129 | Dap3 | 17231 | -0.504 | -0.0419 | No |
| 130 | Smad7 | 17288 | -0.512 | -0.0290 | No |
| 131 | Bnip3l | 17341 | -0.521 | -0.0157 | No |
| 132 | Diablo | 17872 | -0.622 | -0.0254 | No |
| 133 | Tnf | 18056 | -0.686 | -0.0140 | No |
| 134 | Gadd45a | 18381 | -1.070 | 0.0015 | No |