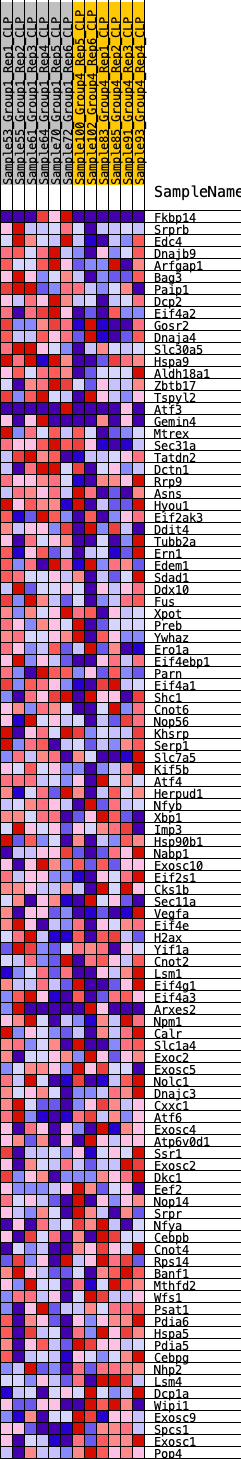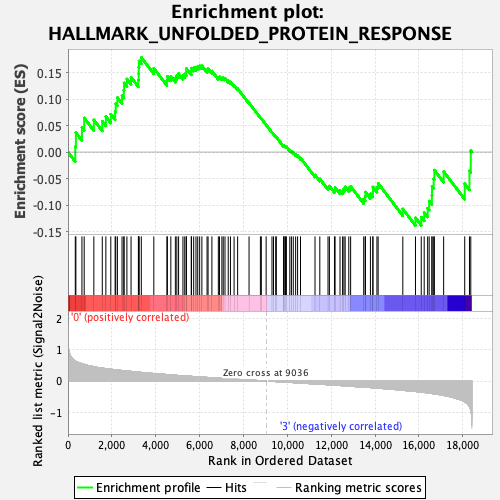
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group1_versus_Group4.Basophil_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_UNFOLDED_PROTEIN_RESPONSE |
| Enrichment Score (ES) | 0.17860553 |
| Normalized Enrichment Score (NES) | 0.70955884 |
| Nominal p-value | 0.81800765 |
| FDR q-value | 0.95970255 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Fkbp14 | 332 | 0.640 | 0.0102 | Yes |
| 2 | Srprb | 359 | 0.628 | 0.0367 | Yes |
| 3 | Edc4 | 635 | 0.558 | 0.0464 | Yes |
| 4 | Dnajb9 | 738 | 0.532 | 0.0644 | Yes |
| 5 | Arfgap1 | 1178 | 0.459 | 0.0607 | Yes |
| 6 | Bag3 | 1563 | 0.414 | 0.0581 | Yes |
| 7 | Paip1 | 1726 | 0.398 | 0.0669 | Yes |
| 8 | Dcp2 | 1951 | 0.380 | 0.0715 | Yes |
| 9 | Eif4a2 | 2148 | 0.362 | 0.0769 | Yes |
| 10 | Gosr2 | 2176 | 0.360 | 0.0914 | Yes |
| 11 | Dnaja4 | 2252 | 0.353 | 0.1029 | Yes |
| 12 | Slc30a5 | 2463 | 0.337 | 0.1064 | Yes |
| 13 | Hspa9 | 2544 | 0.329 | 0.1166 | Yes |
| 14 | Aldh18a1 | 2557 | 0.328 | 0.1305 | Yes |
| 15 | Zbtb17 | 2685 | 0.323 | 0.1379 | Yes |
| 16 | Tspyl2 | 2874 | 0.309 | 0.1413 | Yes |
| 17 | Atf3 | 3208 | 0.285 | 0.1357 | Yes |
| 18 | Gemin4 | 3222 | 0.284 | 0.1476 | Yes |
| 19 | Mtrex | 3226 | 0.284 | 0.1600 | Yes |
| 20 | Sec31a | 3239 | 0.283 | 0.1719 | Yes |
| 21 | Tatdn2 | 3342 | 0.276 | 0.1786 | Yes |
| 22 | Dctn1 | 3912 | 0.239 | 0.1581 | No |
| 23 | Rrp9 | 4509 | 0.208 | 0.1348 | No |
| 24 | Asns | 4530 | 0.207 | 0.1429 | No |
| 25 | Hyou1 | 4688 | 0.198 | 0.1431 | No |
| 26 | Eif2ak3 | 4894 | 0.186 | 0.1401 | No |
| 27 | Ddit4 | 4961 | 0.181 | 0.1445 | No |
| 28 | Tubb2a | 5041 | 0.177 | 0.1481 | No |
| 29 | Ern1 | 5240 | 0.166 | 0.1446 | No |
| 30 | Edem1 | 5322 | 0.163 | 0.1474 | No |
| 31 | Sdad1 | 5391 | 0.160 | 0.1508 | No |
| 32 | Ddx10 | 5395 | 0.159 | 0.1577 | No |
| 33 | Fus | 5619 | 0.151 | 0.1522 | No |
| 34 | Xpot | 5628 | 0.150 | 0.1584 | No |
| 35 | Preb | 5729 | 0.144 | 0.1594 | No |
| 36 | Ywhaz | 5834 | 0.140 | 0.1599 | No |
| 37 | Ero1a | 5913 | 0.136 | 0.1616 | No |
| 38 | Eif4ebp1 | 6001 | 0.133 | 0.1628 | No |
| 39 | Parn | 6103 | 0.127 | 0.1629 | No |
| 40 | Eif4a1 | 6339 | 0.116 | 0.1552 | No |
| 41 | Shc1 | 6385 | 0.113 | 0.1578 | No |
| 42 | Cnot6 | 6560 | 0.106 | 0.1530 | No |
| 43 | Nop56 | 6853 | 0.092 | 0.1411 | No |
| 44 | Khsrp | 6906 | 0.089 | 0.1422 | No |
| 45 | Serp1 | 7015 | 0.083 | 0.1400 | No |
| 46 | Slc7a5 | 7076 | 0.082 | 0.1403 | No |
| 47 | Kif5b | 7157 | 0.079 | 0.1395 | No |
| 48 | Atf4 | 7304 | 0.072 | 0.1347 | No |
| 49 | Herpud1 | 7405 | 0.068 | 0.1323 | No |
| 50 | Nfyb | 7571 | 0.061 | 0.1260 | No |
| 51 | Xbp1 | 7733 | 0.054 | 0.1196 | No |
| 52 | Imp3 | 8253 | 0.033 | 0.0927 | No |
| 53 | Hsp90b1 | 8771 | 0.012 | 0.0650 | No |
| 54 | Nabp1 | 8813 | 0.010 | 0.0632 | No |
| 55 | Exosc10 | 9032 | 0.000 | 0.0513 | No |
| 56 | Eif2s1 | 9299 | -0.007 | 0.0371 | No |
| 57 | Cks1b | 9369 | -0.010 | 0.0337 | No |
| 58 | Sec11a | 9469 | -0.015 | 0.0290 | No |
| 59 | Vegfa | 9496 | -0.016 | 0.0283 | No |
| 60 | Eif4e | 9824 | -0.029 | 0.0117 | No |
| 61 | H2ax | 9841 | -0.029 | 0.0121 | No |
| 62 | Yif1a | 9887 | -0.031 | 0.0110 | No |
| 63 | Cnot2 | 9914 | -0.032 | 0.0110 | No |
| 64 | Lsm1 | 9956 | -0.034 | 0.0103 | No |
| 65 | Eif4g1 | 10112 | -0.040 | 0.0036 | No |
| 66 | Eif4a3 | 10184 | -0.043 | 0.0016 | No |
| 67 | Arxes2 | 10269 | -0.048 | -0.0009 | No |
| 68 | Npm1 | 10377 | -0.053 | -0.0044 | No |
| 69 | Calr | 10466 | -0.057 | -0.0067 | No |
| 70 | Slc1a4 | 10599 | -0.062 | -0.0111 | No |
| 71 | Exoc2 | 11260 | -0.091 | -0.0432 | No |
| 72 | Exosc5 | 11479 | -0.100 | -0.0506 | No |
| 73 | Nolc1 | 11853 | -0.115 | -0.0659 | No |
| 74 | Dnajc3 | 11916 | -0.118 | -0.0641 | No |
| 75 | Cxxc1 | 12147 | -0.128 | -0.0710 | No |
| 76 | Atf6 | 12175 | -0.129 | -0.0667 | No |
| 77 | Exosc4 | 12400 | -0.141 | -0.0727 | No |
| 78 | Atp6v0d1 | 12517 | -0.146 | -0.0726 | No |
| 79 | Ssr1 | 12578 | -0.149 | -0.0693 | No |
| 80 | Exosc2 | 12634 | -0.151 | -0.0655 | No |
| 81 | Dkc1 | 12800 | -0.159 | -0.0675 | No |
| 82 | Eef2 | 12884 | -0.164 | -0.0648 | No |
| 83 | Nop14 | 13480 | -0.193 | -0.0887 | No |
| 84 | Srpr | 13546 | -0.196 | -0.0835 | No |
| 85 | Nfya | 13559 | -0.197 | -0.0755 | No |
| 86 | Cebpb | 13786 | -0.206 | -0.0787 | No |
| 87 | Cnot4 | 13898 | -0.213 | -0.0753 | No |
| 88 | Rps14 | 13899 | -0.213 | -0.0658 | No |
| 89 | Banf1 | 14083 | -0.223 | -0.0660 | No |
| 90 | Mthfd2 | 14138 | -0.225 | -0.0589 | No |
| 91 | Wfs1 | 15263 | -0.293 | -0.1073 | No |
| 92 | Psat1 | 15840 | -0.332 | -0.1241 | No |
| 93 | Pdia6 | 16103 | -0.352 | -0.1228 | No |
| 94 | Hspa5 | 16237 | -0.362 | -0.1140 | No |
| 95 | Pdia5 | 16395 | -0.377 | -0.1059 | No |
| 96 | Cebpg | 16464 | -0.385 | -0.0925 | No |
| 97 | Nhp2 | 16588 | -0.396 | -0.0816 | No |
| 98 | Lsm4 | 16596 | -0.397 | -0.0644 | No |
| 99 | Dcp1a | 16670 | -0.404 | -0.0505 | No |
| 100 | Wipi1 | 16701 | -0.408 | -0.0341 | No |
| 101 | Exosc9 | 17124 | -0.459 | -0.0368 | No |
| 102 | Spcs1 | 18085 | -0.664 | -0.0598 | No |
| 103 | Exosc1 | 18301 | -0.812 | -0.0355 | No |
| 104 | Pop4 | 18361 | -0.931 | 0.0026 | No |