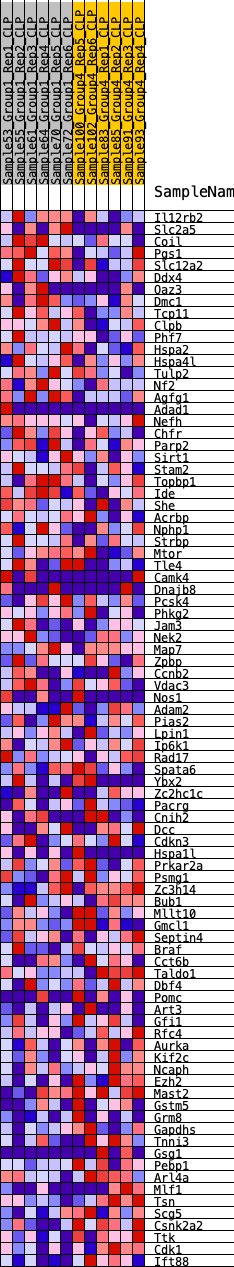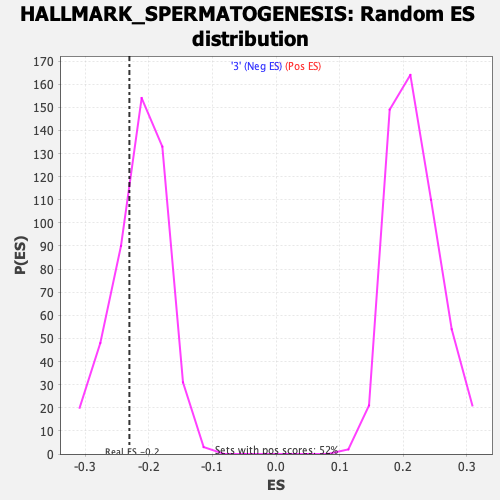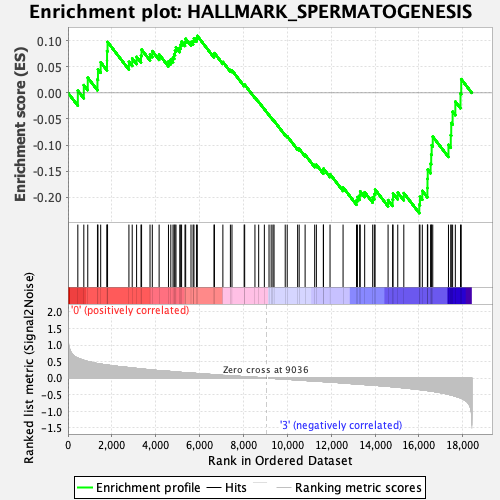
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group1_versus_Group4.Basophil_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_SPERMATOGENESIS |
| Enrichment Score (ES) | -0.23051067 |
| Normalized Enrichment Score (NES) | -1.0761925 |
| Nominal p-value | 0.302714 |
| FDR q-value | 0.58843046 |
| FWER p-Value | 0.989 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Il12rb2 | 448 | 0.601 | 0.0043 | No |
| 2 | Slc2a5 | 721 | 0.535 | 0.0150 | No |
| 3 | Coil | 901 | 0.500 | 0.0291 | No |
| 4 | Pgs1 | 1343 | 0.435 | 0.0258 | No |
| 5 | Slc12a2 | 1366 | 0.431 | 0.0452 | No |
| 6 | Ddx4 | 1490 | 0.421 | 0.0587 | No |
| 7 | Oaz3 | 1778 | 0.391 | 0.0617 | No |
| 8 | Dmc1 | 1781 | 0.391 | 0.0803 | No |
| 9 | Tcp11 | 1803 | 0.388 | 0.0977 | No |
| 10 | Clpb | 2777 | 0.316 | 0.0596 | No |
| 11 | Phf7 | 2927 | 0.304 | 0.0660 | No |
| 12 | Hspa2 | 3127 | 0.290 | 0.0690 | No |
| 13 | Hspa4l | 3328 | 0.276 | 0.0713 | No |
| 14 | Tulp2 | 3354 | 0.274 | 0.0831 | No |
| 15 | Nf2 | 3739 | 0.249 | 0.0740 | No |
| 16 | Agfg1 | 3843 | 0.244 | 0.0801 | No |
| 17 | Adad1 | 4154 | 0.223 | 0.0738 | No |
| 18 | Nefh | 4580 | 0.205 | 0.0604 | No |
| 19 | Chfr | 4687 | 0.198 | 0.0641 | No |
| 20 | Parp2 | 4788 | 0.193 | 0.0678 | No |
| 21 | Sirt1 | 4841 | 0.190 | 0.0741 | No |
| 22 | Stam2 | 4878 | 0.187 | 0.0810 | No |
| 23 | Topbp1 | 4922 | 0.184 | 0.0875 | No |
| 24 | Ide | 5095 | 0.174 | 0.0864 | No |
| 25 | She | 5137 | 0.172 | 0.0924 | No |
| 26 | Acrbp | 5178 | 0.170 | 0.0983 | No |
| 27 | Nphp1 | 5334 | 0.162 | 0.0976 | No |
| 28 | Strbp | 5364 | 0.161 | 0.1037 | No |
| 29 | Mtor | 5607 | 0.152 | 0.0977 | No |
| 30 | Tle4 | 5702 | 0.146 | 0.0996 | No |
| 31 | Camk4 | 5741 | 0.144 | 0.1044 | No |
| 32 | Dnajb8 | 5856 | 0.139 | 0.1048 | No |
| 33 | Pcsk4 | 5894 | 0.137 | 0.1093 | No |
| 34 | Phkg2 | 6654 | 0.101 | 0.0727 | No |
| 35 | Jam3 | 6677 | 0.100 | 0.0763 | No |
| 36 | Nek2 | 7060 | 0.082 | 0.0593 | No |
| 37 | Map7 | 7403 | 0.068 | 0.0439 | No |
| 38 | Zpbp | 7475 | 0.065 | 0.0432 | No |
| 39 | Ccnb2 | 8030 | 0.042 | 0.0149 | No |
| 40 | Vdac3 | 8054 | 0.041 | 0.0157 | No |
| 41 | Nos1 | 8522 | 0.023 | -0.0087 | No |
| 42 | Adam2 | 8692 | 0.015 | -0.0172 | No |
| 43 | Pias2 | 8951 | 0.005 | -0.0311 | No |
| 44 | Lpin1 | 9164 | -0.001 | -0.0426 | No |
| 45 | Ip6k1 | 9261 | -0.005 | -0.0476 | No |
| 46 | Rad17 | 9342 | -0.009 | -0.0515 | No |
| 47 | Spata6 | 9392 | -0.012 | -0.0537 | No |
| 48 | Ybx2 | 9902 | -0.031 | -0.0799 | No |
| 49 | Zc2hc1c | 9993 | -0.035 | -0.0832 | No |
| 50 | Pacrg | 10468 | -0.057 | -0.1063 | No |
| 51 | Cnih2 | 10538 | -0.059 | -0.1073 | No |
| 52 | Dcc | 10809 | -0.071 | -0.1186 | No |
| 53 | Cdkn3 | 11245 | -0.090 | -0.1381 | No |
| 54 | Hspa1l | 11322 | -0.094 | -0.1377 | No |
| 55 | Prkar2a | 11640 | -0.106 | -0.1500 | No |
| 56 | Psmg1 | 11646 | -0.106 | -0.1452 | No |
| 57 | Zc3h14 | 11945 | -0.119 | -0.1558 | No |
| 58 | Bub1 | 12539 | -0.147 | -0.1811 | No |
| 59 | Mllt10 | 13152 | -0.177 | -0.2060 | No |
| 60 | Gmcl1 | 13194 | -0.180 | -0.1997 | No |
| 61 | Septin4 | 13297 | -0.185 | -0.1964 | No |
| 62 | Braf | 13320 | -0.186 | -0.1887 | No |
| 63 | Cct6b | 13524 | -0.196 | -0.1904 | No |
| 64 | Taldo1 | 13889 | -0.212 | -0.2002 | No |
| 65 | Dbf4 | 13964 | -0.217 | -0.1938 | No |
| 66 | Pomc | 13999 | -0.218 | -0.1853 | No |
| 67 | Art3 | 14591 | -0.252 | -0.2055 | No |
| 68 | Gfi1 | 14799 | -0.263 | -0.2042 | No |
| 69 | Rfc4 | 14817 | -0.263 | -0.1926 | No |
| 70 | Aurka | 15032 | -0.274 | -0.1911 | No |
| 71 | Kif2c | 15308 | -0.295 | -0.1920 | No |
| 72 | Ncaph | 16014 | -0.346 | -0.2140 | Yes |
| 73 | Ezh2 | 16044 | -0.347 | -0.1990 | Yes |
| 74 | Mast2 | 16152 | -0.356 | -0.1878 | Yes |
| 75 | Gstm5 | 16383 | -0.376 | -0.1824 | Yes |
| 76 | Grm8 | 16388 | -0.376 | -0.1646 | Yes |
| 77 | Gapdhs | 16399 | -0.377 | -0.1471 | Yes |
| 78 | Tnni3 | 16530 | -0.391 | -0.1355 | Yes |
| 79 | Gsg1 | 16557 | -0.394 | -0.1181 | Yes |
| 80 | Pebp1 | 16579 | -0.395 | -0.1004 | Yes |
| 81 | Arl4a | 16628 | -0.400 | -0.0838 | Yes |
| 82 | Mlf1 | 17344 | -0.488 | -0.0995 | Yes |
| 83 | Tsn | 17450 | -0.507 | -0.0811 | Yes |
| 84 | Scg5 | 17470 | -0.512 | -0.0576 | Yes |
| 85 | Csnk2a2 | 17533 | -0.522 | -0.0360 | Yes |
| 86 | Ttk | 17657 | -0.546 | -0.0166 | Yes |
| 87 | Cdk1 | 17893 | -0.597 | -0.0009 | Yes |
| 88 | Ift88 | 17925 | -0.606 | 0.0264 | Yes |