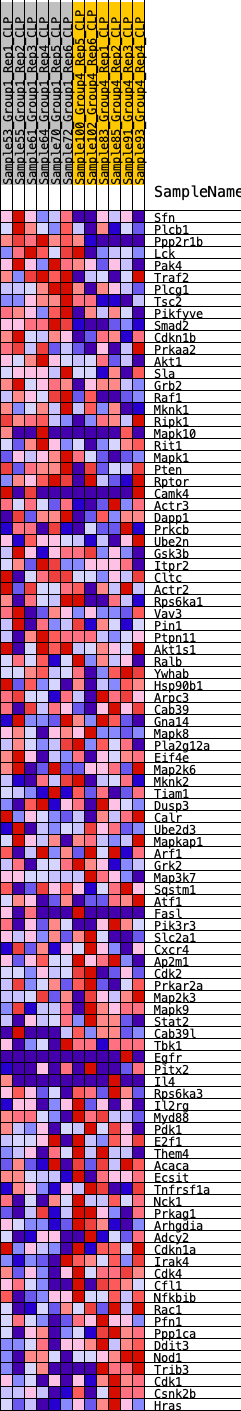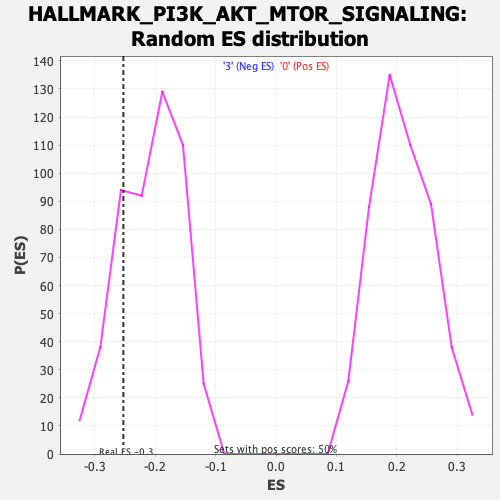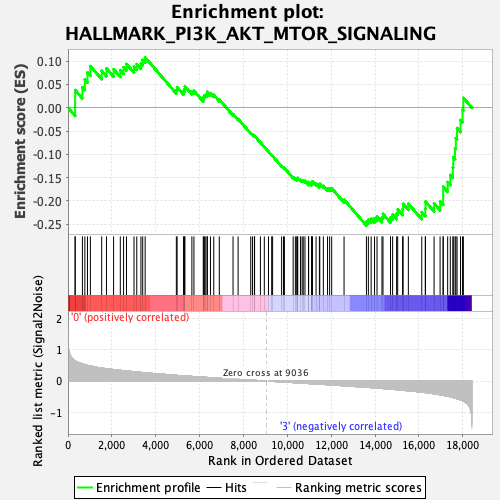
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group1_versus_Group4.Basophil_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_PI3K_AKT_MTOR_SIGNALING |
| Enrichment Score (ES) | -0.25277168 |
| Normalized Enrichment Score (NES) | -1.2192775 |
| Nominal p-value | 0.198 |
| FDR q-value | 0.3900674 |
| FWER p-Value | 0.922 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Sfn | 325 | 0.643 | 0.0100 | No |
| 2 | Plcb1 | 326 | 0.642 | 0.0378 | No |
| 3 | Ppp2r1b | 657 | 0.551 | 0.0435 | No |
| 4 | Lck | 768 | 0.525 | 0.0602 | No |
| 5 | Pak4 | 888 | 0.502 | 0.0754 | No |
| 6 | Traf2 | 1020 | 0.484 | 0.0891 | No |
| 7 | Plcg1 | 1536 | 0.417 | 0.0790 | No |
| 8 | Tsc2 | 1753 | 0.395 | 0.0842 | No |
| 9 | Pikfyve | 2078 | 0.368 | 0.0824 | No |
| 10 | Smad2 | 2384 | 0.343 | 0.0806 | No |
| 11 | Cdkn1b | 2533 | 0.330 | 0.0867 | No |
| 12 | Prkaa2 | 2665 | 0.325 | 0.0936 | No |
| 13 | Akt1 | 3010 | 0.299 | 0.0878 | No |
| 14 | Sla | 3135 | 0.289 | 0.0935 | No |
| 15 | Grb2 | 3325 | 0.277 | 0.0951 | No |
| 16 | Raf1 | 3405 | 0.271 | 0.1025 | No |
| 17 | Mknk1 | 3517 | 0.263 | 0.1078 | No |
| 18 | Ripk1 | 4939 | 0.183 | 0.0380 | No |
| 19 | Mapk10 | 4974 | 0.180 | 0.0440 | No |
| 20 | Rit1 | 5264 | 0.165 | 0.0353 | No |
| 21 | Mapk1 | 5307 | 0.164 | 0.0401 | No |
| 22 | Pten | 5330 | 0.162 | 0.0459 | No |
| 23 | Rptor | 5647 | 0.149 | 0.0351 | No |
| 24 | Camk4 | 5741 | 0.144 | 0.0362 | No |
| 25 | Actr3 | 6167 | 0.124 | 0.0184 | No |
| 26 | Dapp1 | 6185 | 0.123 | 0.0228 | No |
| 27 | Prkcb | 6217 | 0.122 | 0.0263 | No |
| 28 | Ube2n | 6281 | 0.118 | 0.0280 | No |
| 29 | Gsk3b | 6345 | 0.116 | 0.0296 | No |
| 30 | Itpr2 | 6351 | 0.115 | 0.0343 | No |
| 31 | Cltc | 6494 | 0.109 | 0.0312 | No |
| 32 | Actr2 | 6645 | 0.101 | 0.0274 | No |
| 33 | Rps6ka1 | 6897 | 0.090 | 0.0176 | No |
| 34 | Vav3 | 7523 | 0.062 | -0.0139 | No |
| 35 | Pin1 | 7754 | 0.053 | -0.0241 | No |
| 36 | Ptpn11 | 8330 | 0.030 | -0.0542 | No |
| 37 | Akt1s1 | 8409 | 0.027 | -0.0573 | No |
| 38 | Ralb | 8497 | 0.024 | -0.0610 | No |
| 39 | Ywhab | 8506 | 0.024 | -0.0605 | No |
| 40 | Hsp90b1 | 8771 | 0.012 | -0.0744 | No |
| 41 | Arpc3 | 8949 | 0.005 | -0.0839 | No |
| 42 | Cab39 | 9140 | -0.000 | -0.0942 | No |
| 43 | Gna14 | 9285 | -0.006 | -0.1018 | No |
| 44 | Mapk8 | 9327 | -0.008 | -0.1037 | No |
| 45 | Pla2g12a | 9738 | -0.025 | -0.1250 | No |
| 46 | Eif4e | 9824 | -0.029 | -0.1284 | No |
| 47 | Map2k6 | 9865 | -0.030 | -0.1293 | No |
| 48 | Mknk2 | 10264 | -0.047 | -0.1490 | No |
| 49 | Tiam1 | 10365 | -0.052 | -0.1522 | No |
| 50 | Dusp3 | 10425 | -0.055 | -0.1531 | No |
| 51 | Calr | 10466 | -0.057 | -0.1528 | No |
| 52 | Ube2d3 | 10470 | -0.057 | -0.1505 | No |
| 53 | Mapkap1 | 10597 | -0.062 | -0.1547 | No |
| 54 | Arf1 | 10672 | -0.064 | -0.1560 | No |
| 55 | Grk2 | 10723 | -0.067 | -0.1558 | No |
| 56 | Map3k7 | 10802 | -0.071 | -0.1570 | No |
| 57 | Sqstm1 | 10966 | -0.078 | -0.1626 | No |
| 58 | Atf1 | 10968 | -0.078 | -0.1593 | No |
| 59 | Fasl | 11103 | -0.083 | -0.1630 | No |
| 60 | Pik3r3 | 11110 | -0.083 | -0.1597 | No |
| 61 | Slc2a1 | 11145 | -0.085 | -0.1579 | No |
| 62 | Cxcr4 | 11308 | -0.093 | -0.1628 | No |
| 63 | Ap2m1 | 11459 | -0.099 | -0.1667 | No |
| 64 | Cdk2 | 11478 | -0.100 | -0.1633 | No |
| 65 | Prkar2a | 11640 | -0.106 | -0.1675 | No |
| 66 | Map2k3 | 11831 | -0.114 | -0.1730 | No |
| 67 | Mapk9 | 11926 | -0.118 | -0.1730 | No |
| 68 | Stat2 | 12020 | -0.122 | -0.1728 | No |
| 69 | Cab39l | 12584 | -0.149 | -0.1971 | No |
| 70 | Tbk1 | 13604 | -0.200 | -0.2441 | Yes |
| 71 | Egfr | 13692 | -0.205 | -0.2400 | Yes |
| 72 | Pitx2 | 13819 | -0.208 | -0.2379 | Yes |
| 73 | Il4 | 13974 | -0.218 | -0.2369 | Yes |
| 74 | Rps6ka3 | 14087 | -0.223 | -0.2334 | Yes |
| 75 | Il2rg | 14312 | -0.236 | -0.2355 | Yes |
| 76 | Myd88 | 14359 | -0.238 | -0.2277 | Yes |
| 77 | Pdk1 | 14702 | -0.256 | -0.2353 | Yes |
| 78 | E2f1 | 14798 | -0.263 | -0.2291 | Yes |
| 79 | Them4 | 14981 | -0.272 | -0.2273 | Yes |
| 80 | Acaca | 15029 | -0.274 | -0.2181 | Yes |
| 81 | Ecsit | 15253 | -0.292 | -0.2176 | Yes |
| 82 | Tnfrsf1a | 15276 | -0.294 | -0.2061 | Yes |
| 83 | Nck1 | 15515 | -0.308 | -0.2058 | Yes |
| 84 | Prkag1 | 16127 | -0.354 | -0.2239 | Yes |
| 85 | Arhgdia | 16287 | -0.368 | -0.2167 | Yes |
| 86 | Adcy2 | 16297 | -0.368 | -0.2013 | Yes |
| 87 | Cdkn1a | 16688 | -0.407 | -0.2050 | Yes |
| 88 | Irak4 | 16962 | -0.437 | -0.2010 | Yes |
| 89 | Cdk4 | 17101 | -0.457 | -0.1888 | Yes |
| 90 | Cfl1 | 17103 | -0.457 | -0.1691 | Yes |
| 91 | Nfkbib | 17307 | -0.481 | -0.1594 | Yes |
| 92 | Rac1 | 17427 | -0.503 | -0.1442 | Yes |
| 93 | Pfn1 | 17546 | -0.525 | -0.1280 | Yes |
| 94 | Ppp1ca | 17562 | -0.527 | -0.1061 | Yes |
| 95 | Ddit3 | 17644 | -0.543 | -0.0870 | Yes |
| 96 | Nod1 | 17681 | -0.552 | -0.0651 | Yes |
| 97 | Trib3 | 17735 | -0.563 | -0.0437 | Yes |
| 98 | Cdk1 | 17893 | -0.597 | -0.0265 | Yes |
| 99 | Csnk2b | 17985 | -0.625 | -0.0045 | Yes |
| 100 | Hras | 18023 | -0.638 | 0.0210 | Yes |