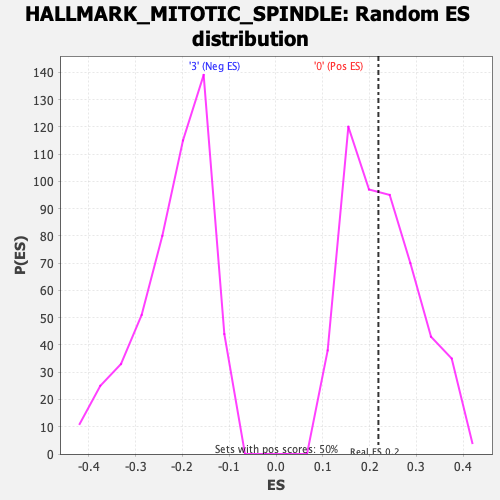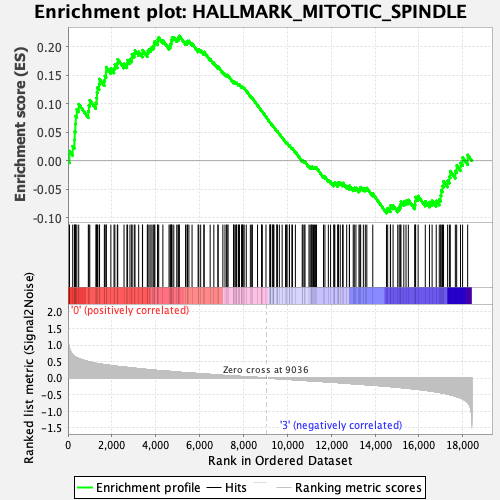
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group1_versus_Group4.Basophil_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_MITOTIC_SPINDLE |
| Enrichment Score (ES) | 0.21879928 |
| Normalized Enrichment Score (NES) | 0.9599337 |
| Nominal p-value | 0.49601594 |
| FDR q-value | 0.8645343 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Sptan1 | 68 | 0.895 | 0.0172 | Yes |
| 2 | Stk38l | 214 | 0.702 | 0.0257 | Yes |
| 3 | Arhgef12 | 292 | 0.658 | 0.0368 | Yes |
| 4 | Sun2 | 305 | 0.651 | 0.0514 | Yes |
| 5 | Gsn | 337 | 0.636 | 0.0646 | Yes |
| 6 | Als2 | 352 | 0.630 | 0.0785 | Yes |
| 7 | Dynll2 | 402 | 0.614 | 0.0902 | Yes |
| 8 | Kif3b | 488 | 0.589 | 0.0993 | Yes |
| 9 | Sptbn1 | 926 | 0.496 | 0.0869 | Yes |
| 10 | Pdlim5 | 951 | 0.492 | 0.0971 | Yes |
| 11 | Mid1 | 993 | 0.487 | 0.1062 | Yes |
| 12 | Numa1 | 1271 | 0.445 | 0.1015 | Yes |
| 13 | Tubd1 | 1301 | 0.441 | 0.1102 | Yes |
| 14 | Pkd2 | 1320 | 0.439 | 0.1195 | Yes |
| 15 | Arhgap29 | 1342 | 0.435 | 0.1285 | Yes |
| 16 | Ppp4r2 | 1418 | 0.426 | 0.1343 | Yes |
| 17 | Arf6 | 1438 | 0.425 | 0.1432 | Yes |
| 18 | Arhgap4 | 1657 | 0.403 | 0.1407 | Yes |
| 19 | Myh9 | 1683 | 0.402 | 0.1487 | Yes |
| 20 | Ophn1 | 1739 | 0.397 | 0.1550 | Yes |
| 21 | Tubgcp6 | 1742 | 0.397 | 0.1641 | Yes |
| 22 | Fgd4 | 1954 | 0.379 | 0.1614 | Yes |
| 23 | Cep131 | 2098 | 0.365 | 0.1621 | Yes |
| 24 | Fgd6 | 2136 | 0.363 | 0.1686 | Yes |
| 25 | Cttn | 2248 | 0.353 | 0.1707 | Yes |
| 26 | Arfip2 | 2267 | 0.352 | 0.1780 | Yes |
| 27 | Rasa1 | 2554 | 0.328 | 0.1699 | Yes |
| 28 | Cep192 | 2686 | 0.323 | 0.1703 | Yes |
| 29 | Rasa2 | 2716 | 0.320 | 0.1762 | Yes |
| 30 | Arhgap10 | 2825 | 0.313 | 0.1776 | Yes |
| 31 | Epb41 | 2909 | 0.306 | 0.1802 | Yes |
| 32 | Nin | 2917 | 0.305 | 0.1869 | Yes |
| 33 | Flna | 3020 | 0.299 | 0.1883 | Yes |
| 34 | Tbcd | 3054 | 0.296 | 0.1934 | Yes |
| 35 | Gemin4 | 3222 | 0.284 | 0.1909 | Yes |
| 36 | Trio | 3393 | 0.272 | 0.1879 | Yes |
| 37 | Itsn1 | 3400 | 0.271 | 0.1939 | Yes |
| 38 | Wasl | 3615 | 0.256 | 0.1881 | Yes |
| 39 | Vcl | 3643 | 0.254 | 0.1926 | Yes |
| 40 | Arhgef2 | 3697 | 0.251 | 0.1955 | Yes |
| 41 | Bcr | 3777 | 0.246 | 0.1970 | Yes |
| 42 | Hook3 | 3833 | 0.246 | 0.1997 | Yes |
| 43 | Lrpprc | 3910 | 0.239 | 0.2011 | Yes |
| 44 | Tubgcp2 | 3927 | 0.238 | 0.2058 | Yes |
| 45 | Cdk5rap2 | 3960 | 0.236 | 0.2095 | Yes |
| 46 | Bin1 | 4080 | 0.227 | 0.2083 | Yes |
| 47 | Abr | 4097 | 0.226 | 0.2127 | Yes |
| 48 | Notch2 | 4135 | 0.224 | 0.2159 | Yes |
| 49 | Rab3gap1 | 4325 | 0.217 | 0.2106 | Yes |
| 50 | Lats1 | 4606 | 0.203 | 0.2000 | Yes |
| 51 | Arhgef7 | 4657 | 0.200 | 0.2019 | Yes |
| 52 | Farp1 | 4683 | 0.198 | 0.2052 | Yes |
| 53 | Cntrob | 4711 | 0.197 | 0.2083 | Yes |
| 54 | Flnb | 4722 | 0.196 | 0.2124 | Yes |
| 55 | Arfgef1 | 4743 | 0.195 | 0.2158 | Yes |
| 56 | Ranbp9 | 4822 | 0.191 | 0.2160 | Yes |
| 57 | Akap13 | 4958 | 0.181 | 0.2128 | Yes |
| 58 | Lmnb1 | 5017 | 0.178 | 0.2138 | Yes |
| 59 | Capzb | 5045 | 0.177 | 0.2165 | Yes |
| 60 | Actn4 | 5078 | 0.175 | 0.2188 | Yes |
| 61 | Stau1 | 5359 | 0.161 | 0.2072 | No |
| 62 | Pafah1b1 | 5426 | 0.157 | 0.2072 | No |
| 63 | Dync1h1 | 5445 | 0.156 | 0.2099 | No |
| 64 | Shroom2 | 5511 | 0.154 | 0.2099 | No |
| 65 | Ckap5 | 5655 | 0.148 | 0.2055 | No |
| 66 | Smc1a | 5931 | 0.136 | 0.1936 | No |
| 67 | Mid1ip1 | 5962 | 0.135 | 0.1951 | No |
| 68 | Mark4 | 6048 | 0.130 | 0.1935 | No |
| 69 | Cep250 | 6192 | 0.123 | 0.1885 | No |
| 70 | Smc3 | 6207 | 0.122 | 0.1906 | No |
| 71 | Pcnt | 6480 | 0.109 | 0.1782 | No |
| 72 | Map3k11 | 6648 | 0.101 | 0.1714 | No |
| 73 | Kif3c | 6836 | 0.093 | 0.1633 | No |
| 74 | Kifap3 | 6841 | 0.093 | 0.1653 | No |
| 75 | Nek2 | 7060 | 0.082 | 0.1552 | No |
| 76 | Kif5b | 7157 | 0.079 | 0.1518 | No |
| 77 | Pcm1 | 7234 | 0.075 | 0.1494 | No |
| 78 | Synpo | 7249 | 0.074 | 0.1504 | No |
| 79 | Nck2 | 7297 | 0.073 | 0.1495 | No |
| 80 | Cntrl | 7545 | 0.062 | 0.1374 | No |
| 81 | Myo1e | 7553 | 0.061 | 0.1384 | No |
| 82 | Palld | 7572 | 0.061 | 0.1388 | No |
| 83 | Rapgef6 | 7645 | 0.058 | 0.1362 | No |
| 84 | Tlk1 | 7674 | 0.057 | 0.1360 | No |
| 85 | Pcgf5 | 7693 | 0.056 | 0.1364 | No |
| 86 | Dlg1 | 7771 | 0.053 | 0.1334 | No |
| 87 | Csnk1d | 7795 | 0.052 | 0.1333 | No |
| 88 | Ect2 | 7815 | 0.051 | 0.1335 | No |
| 89 | Rfc1 | 7914 | 0.046 | 0.1292 | No |
| 90 | Cep72 | 7923 | 0.046 | 0.1298 | No |
| 91 | Ywhae | 7952 | 0.045 | 0.1293 | No |
| 92 | Prex1 | 7984 | 0.044 | 0.1286 | No |
| 93 | Ccnb2 | 8030 | 0.042 | 0.1272 | No |
| 94 | Katna1 | 8126 | 0.038 | 0.1228 | No |
| 95 | Tsc1 | 8312 | 0.030 | 0.1134 | No |
| 96 | Apc | 8376 | 0.028 | 0.1106 | No |
| 97 | Klc1 | 8404 | 0.027 | 0.1098 | No |
| 98 | Epb41l2 | 8640 | 0.018 | 0.0973 | No |
| 99 | Arl8a | 8824 | 0.010 | 0.0875 | No |
| 100 | Myo9b | 8835 | 0.009 | 0.0871 | No |
| 101 | Rock1 | 8853 | 0.009 | 0.0864 | No |
| 102 | Clip1 | 9025 | 0.001 | 0.0770 | No |
| 103 | Net1 | 9206 | -0.003 | 0.0672 | No |
| 104 | Taok2 | 9212 | -0.003 | 0.0670 | No |
| 105 | Cenpf | 9221 | -0.004 | 0.0667 | No |
| 106 | Clasp1 | 9232 | -0.004 | 0.0662 | No |
| 107 | Abi1 | 9313 | -0.008 | 0.0620 | No |
| 108 | Cyth2 | 9348 | -0.009 | 0.0604 | No |
| 109 | Nf1 | 9359 | -0.010 | 0.0600 | No |
| 110 | Rhof | 9387 | -0.011 | 0.0588 | No |
| 111 | Wasf1 | 9522 | -0.017 | 0.0518 | No |
| 112 | Prc1 | 9535 | -0.017 | 0.0516 | No |
| 113 | Espl1 | 9538 | -0.017 | 0.0519 | No |
| 114 | Kif11 | 9635 | -0.021 | 0.0471 | No |
| 115 | Rabgap1 | 9762 | -0.026 | 0.0408 | No |
| 116 | Mapre1 | 9917 | -0.032 | 0.0331 | No |
| 117 | Dst | 9969 | -0.034 | 0.0311 | No |
| 118 | Ezr | 9985 | -0.035 | 0.0311 | No |
| 119 | Atg4b | 10092 | -0.039 | 0.0262 | No |
| 120 | Tubgcp3 | 10094 | -0.039 | 0.0270 | No |
| 121 | Dlgap5 | 10207 | -0.045 | 0.0219 | No |
| 122 | Fscn1 | 10231 | -0.046 | 0.0218 | No |
| 123 | Tiam1 | 10365 | -0.052 | 0.0157 | No |
| 124 | Sos1 | 10677 | -0.065 | 0.0001 | No |
| 125 | Ralbp1 | 10709 | -0.067 | -0.0000 | No |
| 126 | Incenp | 10765 | -0.069 | -0.0014 | No |
| 127 | Nedd9 | 10804 | -0.071 | -0.0019 | No |
| 128 | Kif20b | 10986 | -0.078 | -0.0100 | No |
| 129 | Dock2 | 11038 | -0.081 | -0.0109 | No |
| 130 | Abl1 | 11087 | -0.082 | -0.0116 | No |
| 131 | Kif1b | 11120 | -0.084 | -0.0114 | No |
| 132 | Plk1 | 11132 | -0.084 | -0.0100 | No |
| 133 | Kif4 | 11191 | -0.087 | -0.0112 | No |
| 134 | Top2a | 11241 | -0.090 | -0.0118 | No |
| 135 | Wasf2 | 11279 | -0.091 | -0.0117 | No |
| 136 | Sass6 | 11329 | -0.094 | -0.0121 | No |
| 137 | Cdc42bpa | 11647 | -0.106 | -0.0271 | No |
| 138 | Arhgap5 | 11712 | -0.109 | -0.0280 | No |
| 139 | Bcl2l11 | 11868 | -0.116 | -0.0339 | No |
| 140 | Cdc42ep4 | 11970 | -0.120 | -0.0366 | No |
| 141 | Pxn | 12114 | -0.127 | -0.0415 | No |
| 142 | Kif22 | 12138 | -0.127 | -0.0398 | No |
| 143 | Sac3d1 | 12163 | -0.128 | -0.0381 | No |
| 144 | Cd2ap | 12294 | -0.134 | -0.0421 | No |
| 145 | Kif23 | 12320 | -0.136 | -0.0403 | No |
| 146 | Cenpe | 12328 | -0.136 | -0.0375 | No |
| 147 | Myh10 | 12411 | -0.141 | -0.0387 | No |
| 148 | Arhgef11 | 12520 | -0.146 | -0.0412 | No |
| 149 | Bub1 | 12539 | -0.147 | -0.0387 | No |
| 150 | Rasal2 | 12704 | -0.154 | -0.0441 | No |
| 151 | Anln | 12825 | -0.161 | -0.0469 | No |
| 152 | Arhgef3 | 12833 | -0.161 | -0.0436 | No |
| 153 | Cdc42 | 13002 | -0.170 | -0.0488 | No |
| 154 | Pif1 | 13050 | -0.172 | -0.0474 | No |
| 155 | Tubgcp5 | 13125 | -0.176 | -0.0473 | No |
| 156 | Tpx2 | 13264 | -0.183 | -0.0506 | No |
| 157 | Rictor | 13307 | -0.185 | -0.0486 | No |
| 158 | Rhot2 | 13341 | -0.187 | -0.0460 | No |
| 159 | Smc4 | 13464 | -0.192 | -0.0482 | No |
| 160 | Plekhg2 | 13553 | -0.197 | -0.0485 | No |
| 161 | Septin9 | 13622 | -0.201 | -0.0475 | No |
| 162 | Cdc27 | 13893 | -0.213 | -0.0574 | No |
| 163 | Alms1 | 14518 | -0.247 | -0.0858 | No |
| 164 | Ssh2 | 14573 | -0.250 | -0.0830 | No |
| 165 | Kptn | 14695 | -0.256 | -0.0836 | No |
| 166 | Llgl1 | 14704 | -0.256 | -0.0781 | No |
| 167 | Katnb1 | 14819 | -0.264 | -0.0782 | No |
| 168 | Aurka | 15032 | -0.274 | -0.0834 | No |
| 169 | Brca2 | 15097 | -0.279 | -0.0804 | No |
| 170 | Ndc80 | 15149 | -0.283 | -0.0765 | No |
| 171 | Nusap1 | 15179 | -0.286 | -0.0715 | No |
| 172 | Kif2c | 15308 | -0.295 | -0.0716 | No |
| 173 | Racgap1 | 15409 | -0.304 | -0.0700 | No |
| 174 | Nck1 | 15515 | -0.308 | -0.0685 | No |
| 175 | Kif15 | 15806 | -0.329 | -0.0767 | No |
| 176 | Cep57 | 15829 | -0.331 | -0.0702 | No |
| 177 | Map1s | 15847 | -0.332 | -0.0634 | No |
| 178 | Marcks | 15961 | -0.342 | -0.0616 | No |
| 179 | Arhgdia | 16287 | -0.368 | -0.0708 | No |
| 180 | Cenpj | 16477 | -0.386 | -0.0722 | No |
| 181 | Arhgap27 | 16598 | -0.397 | -0.0695 | No |
| 182 | Fbxo5 | 16786 | -0.415 | -0.0700 | No |
| 183 | Birc5 | 16925 | -0.432 | -0.0675 | No |
| 184 | Sorbs2 | 16989 | -0.442 | -0.0606 | No |
| 185 | Kntc1 | 17012 | -0.445 | -0.0514 | No |
| 186 | Arap3 | 17072 | -0.453 | -0.0441 | No |
| 187 | Uxt | 17119 | -0.458 | -0.0359 | No |
| 188 | Clip2 | 17305 | -0.481 | -0.0348 | No |
| 189 | Ccdc88a | 17387 | -0.496 | -0.0276 | No |
| 190 | Tuba4a | 17426 | -0.503 | -0.0180 | No |
| 191 | Ttk | 17657 | -0.546 | -0.0178 | No |
| 192 | Rapgef5 | 17718 | -0.559 | -0.0080 | No |
| 193 | Cdk1 | 17893 | -0.597 | -0.0036 | No |
| 194 | Hdac6 | 17988 | -0.625 | 0.0058 | No |
| 195 | Dock4 | 18219 | -0.734 | 0.0104 | No |