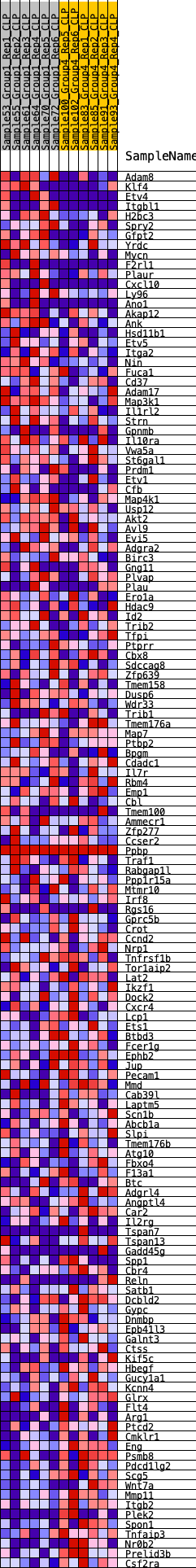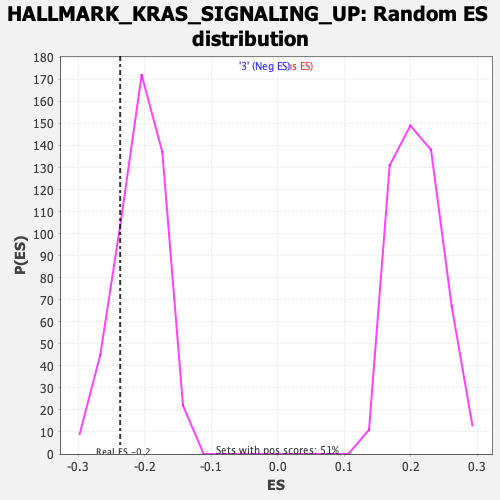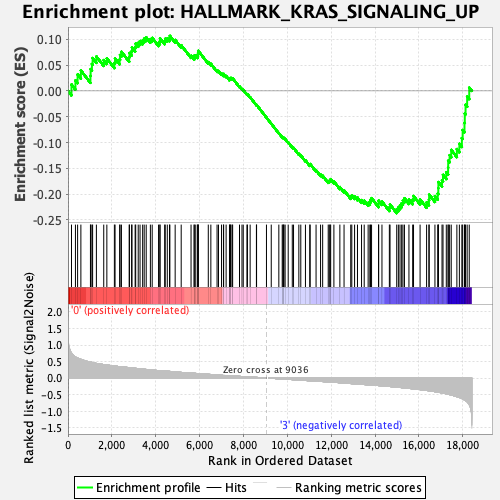
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group1_versus_Group4.Basophil_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_KRAS_SIGNALING_UP |
| Enrichment Score (ES) | -0.23674546 |
| Normalized Enrichment Score (NES) | -1.1384075 |
| Nominal p-value | 0.17718941 |
| FDR q-value | 0.4755741 |
| FWER p-Value | 0.969 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Adam8 | 159 | 0.753 | 0.0123 | No |
| 2 | Klf4 | 339 | 0.635 | 0.0203 | No |
| 3 | Etv4 | 438 | 0.602 | 0.0318 | No |
| 4 | Itgbl1 | 587 | 0.565 | 0.0394 | No |
| 5 | H2bc3 | 1023 | 0.484 | 0.0291 | No |
| 6 | Spry2 | 1032 | 0.482 | 0.0422 | No |
| 7 | Gfpt2 | 1091 | 0.472 | 0.0522 | No |
| 8 | Yrdc | 1122 | 0.467 | 0.0636 | No |
| 9 | Mycn | 1294 | 0.442 | 0.0666 | No |
| 10 | F2rl1 | 1629 | 0.406 | 0.0597 | No |
| 11 | Plaur | 1771 | 0.392 | 0.0629 | No |
| 12 | Cxcl10 | 2114 | 0.364 | 0.0543 | No |
| 13 | Ly96 | 2145 | 0.362 | 0.0628 | No |
| 14 | Ano1 | 2355 | 0.344 | 0.0610 | No |
| 15 | Akap12 | 2373 | 0.344 | 0.0697 | No |
| 16 | Ank | 2436 | 0.339 | 0.0757 | No |
| 17 | Hsd11b1 | 2789 | 0.315 | 0.0653 | No |
| 18 | Etv5 | 2802 | 0.314 | 0.0734 | No |
| 19 | Itga2 | 2899 | 0.306 | 0.0767 | No |
| 20 | Nin | 2917 | 0.305 | 0.0843 | No |
| 21 | Fuca1 | 3061 | 0.295 | 0.0847 | No |
| 22 | Cd37 | 3086 | 0.293 | 0.0916 | No |
| 23 | Adam17 | 3201 | 0.285 | 0.0933 | No |
| 24 | Map3k1 | 3284 | 0.280 | 0.0966 | No |
| 25 | Il1rl2 | 3403 | 0.271 | 0.0977 | No |
| 26 | Strn | 3473 | 0.265 | 0.1014 | No |
| 27 | Gpnmb | 3565 | 0.260 | 0.1037 | No |
| 28 | Il10ra | 3755 | 0.248 | 0.1002 | No |
| 29 | Vwa5a | 3838 | 0.245 | 0.1026 | No |
| 30 | St6gal1 | 4128 | 0.225 | 0.0930 | No |
| 31 | Prdm1 | 4187 | 0.222 | 0.0961 | No |
| 32 | Etv1 | 4196 | 0.222 | 0.1018 | No |
| 33 | Cfb | 4408 | 0.214 | 0.0963 | No |
| 34 | Map4k1 | 4431 | 0.213 | 0.1010 | No |
| 35 | Usp12 | 4526 | 0.207 | 0.1017 | No |
| 36 | Akt2 | 4627 | 0.202 | 0.1018 | No |
| 37 | Avl9 | 4644 | 0.201 | 0.1066 | No |
| 38 | Evi5 | 4885 | 0.186 | 0.0986 | No |
| 39 | Adgra2 | 5160 | 0.170 | 0.0884 | No |
| 40 | Birc3 | 5609 | 0.151 | 0.0681 | No |
| 41 | Gng11 | 5752 | 0.143 | 0.0643 | No |
| 42 | Plvap | 5753 | 0.143 | 0.0683 | No |
| 43 | Plau | 5824 | 0.140 | 0.0684 | No |
| 44 | Ero1a | 5913 | 0.136 | 0.0674 | No |
| 45 | Hdac9 | 5919 | 0.136 | 0.0709 | No |
| 46 | Id2 | 5933 | 0.135 | 0.0740 | No |
| 47 | Trib2 | 5937 | 0.135 | 0.0776 | No |
| 48 | Tfpi | 6395 | 0.113 | 0.0557 | No |
| 49 | Ptprr | 6512 | 0.108 | 0.0524 | No |
| 50 | Cbx8 | 6795 | 0.095 | 0.0396 | No |
| 51 | Sdccag8 | 6860 | 0.092 | 0.0387 | No |
| 52 | Zfp639 | 6999 | 0.084 | 0.0335 | No |
| 53 | Tmem158 | 7087 | 0.081 | 0.0310 | No |
| 54 | Dusp6 | 7095 | 0.081 | 0.0329 | No |
| 55 | Wdr33 | 7208 | 0.077 | 0.0289 | No |
| 56 | Trib1 | 7361 | 0.070 | 0.0225 | No |
| 57 | Tmem176a | 7373 | 0.070 | 0.0238 | No |
| 58 | Map7 | 7403 | 0.068 | 0.0242 | No |
| 59 | Ptbp2 | 7420 | 0.068 | 0.0252 | No |
| 60 | Bpgm | 7480 | 0.065 | 0.0238 | No |
| 61 | Cdadc1 | 7505 | 0.063 | 0.0242 | No |
| 62 | Il7r | 7818 | 0.051 | 0.0086 | No |
| 63 | Rbm4 | 7931 | 0.046 | 0.0037 | No |
| 64 | Emp1 | 7989 | 0.044 | 0.0018 | No |
| 65 | Cbl | 8158 | 0.037 | -0.0063 | No |
| 66 | Tmem100 | 8180 | 0.037 | -0.0065 | No |
| 67 | Ammecr1 | 8303 | 0.030 | -0.0123 | No |
| 68 | Zfp277 | 8589 | 0.021 | -0.0273 | No |
| 69 | Ccser2 | 8596 | 0.020 | -0.0271 | No |
| 70 | Ppbp | 9047 | 0.000 | -0.0517 | No |
| 71 | Traf1 | 9265 | -0.006 | -0.0634 | No |
| 72 | Rabgap1l | 9611 | -0.020 | -0.0818 | No |
| 73 | Ppp1r15a | 9765 | -0.026 | -0.0894 | No |
| 74 | Mtmr10 | 9799 | -0.028 | -0.0904 | No |
| 75 | Irf8 | 9822 | -0.029 | -0.0908 | No |
| 76 | Rgs16 | 9844 | -0.030 | -0.0912 | No |
| 77 | Gprc5b | 9897 | -0.031 | -0.0931 | No |
| 78 | Crot | 10038 | -0.037 | -0.0998 | No |
| 79 | Ccnd2 | 10224 | -0.046 | -0.1086 | No |
| 80 | Nrp1 | 10281 | -0.048 | -0.1103 | No |
| 81 | Tnfrsf1b | 10523 | -0.059 | -0.1219 | No |
| 82 | Tor1aip2 | 10612 | -0.062 | -0.1250 | No |
| 83 | Lat2 | 10824 | -0.072 | -0.1345 | No |
| 84 | Ikzf1 | 11022 | -0.080 | -0.1431 | No |
| 85 | Dock2 | 11038 | -0.081 | -0.1416 | No |
| 86 | Cxcr4 | 11308 | -0.093 | -0.1538 | No |
| 87 | Lcp1 | 11513 | -0.102 | -0.1621 | No |
| 88 | Ets1 | 11609 | -0.104 | -0.1644 | No |
| 89 | Btbd3 | 11862 | -0.115 | -0.1749 | No |
| 90 | Fcer1g | 11909 | -0.118 | -0.1742 | No |
| 91 | Ephb2 | 11946 | -0.119 | -0.1728 | No |
| 92 | Jup | 11974 | -0.121 | -0.1709 | No |
| 93 | Pecam1 | 12121 | -0.127 | -0.1754 | No |
| 94 | Mmd | 12393 | -0.140 | -0.1863 | No |
| 95 | Cab39l | 12584 | -0.149 | -0.1925 | No |
| 96 | Laptm5 | 12889 | -0.165 | -0.2046 | No |
| 97 | Scn1b | 12937 | -0.167 | -0.2025 | No |
| 98 | Abcb1a | 13064 | -0.173 | -0.2045 | No |
| 99 | Slpi | 13196 | -0.180 | -0.2067 | No |
| 100 | Tmem176b | 13381 | -0.189 | -0.2115 | No |
| 101 | Atg10 | 13503 | -0.194 | -0.2127 | No |
| 102 | Fbxo4 | 13682 | -0.205 | -0.2167 | No |
| 103 | F13a1 | 13760 | -0.205 | -0.2152 | No |
| 104 | Btc | 13789 | -0.207 | -0.2109 | No |
| 105 | Adgrl4 | 13838 | -0.209 | -0.2077 | No |
| 106 | Angptl4 | 14159 | -0.226 | -0.2189 | No |
| 107 | Car2 | 14160 | -0.226 | -0.2126 | No |
| 108 | Il2rg | 14312 | -0.236 | -0.2143 | No |
| 109 | Tspan7 | 14649 | -0.254 | -0.2256 | No |
| 110 | Tspan13 | 14680 | -0.255 | -0.2201 | No |
| 111 | Gadd45g | 14985 | -0.272 | -0.2291 | Yes |
| 112 | Spp1 | 15060 | -0.276 | -0.2255 | Yes |
| 113 | Cbr4 | 15134 | -0.282 | -0.2216 | Yes |
| 114 | Reln | 15210 | -0.288 | -0.2176 | Yes |
| 115 | Satb1 | 15268 | -0.293 | -0.2125 | Yes |
| 116 | Dcbld2 | 15337 | -0.298 | -0.2079 | Yes |
| 117 | Gypc | 15542 | -0.311 | -0.2104 | Yes |
| 118 | Dnmbp | 15712 | -0.324 | -0.2106 | Yes |
| 119 | Epb41l3 | 15751 | -0.326 | -0.2036 | Yes |
| 120 | Galnt3 | 16050 | -0.348 | -0.2102 | Yes |
| 121 | Ctss | 16347 | -0.371 | -0.2160 | Yes |
| 122 | Kif5c | 16454 | -0.383 | -0.2111 | Yes |
| 123 | Hbegf | 16463 | -0.385 | -0.2008 | Yes |
| 124 | Gucy1a1 | 16727 | -0.410 | -0.2037 | Yes |
| 125 | Kcnn4 | 16855 | -0.423 | -0.1988 | Yes |
| 126 | Glrx | 16881 | -0.426 | -0.1883 | Yes |
| 127 | Flt4 | 16885 | -0.426 | -0.1766 | Yes |
| 128 | Arg1 | 17050 | -0.450 | -0.1730 | Yes |
| 129 | Ptcd2 | 17095 | -0.457 | -0.1626 | Yes |
| 130 | Cmklr1 | 17245 | -0.474 | -0.1575 | Yes |
| 131 | Eng | 17328 | -0.485 | -0.1485 | Yes |
| 132 | Psmb8 | 17332 | -0.486 | -0.1350 | Yes |
| 133 | Pdcd1lg2 | 17396 | -0.498 | -0.1246 | Yes |
| 134 | Scg5 | 17470 | -0.512 | -0.1143 | Yes |
| 135 | Wnt7a | 17727 | -0.561 | -0.1126 | Yes |
| 136 | Mmp11 | 17841 | -0.586 | -0.1024 | Yes |
| 137 | Itgb2 | 17955 | -0.617 | -0.0914 | Yes |
| 138 | Plek2 | 17991 | -0.626 | -0.0758 | Yes |
| 139 | Spon1 | 18074 | -0.661 | -0.0618 | Yes |
| 140 | Tnfaip3 | 18088 | -0.664 | -0.0440 | Yes |
| 141 | Nr0b2 | 18123 | -0.678 | -0.0269 | Yes |
| 142 | Prelid3b | 18196 | -0.715 | -0.0108 | Yes |
| 143 | Csf2ra | 18291 | -0.801 | 0.0064 | Yes |