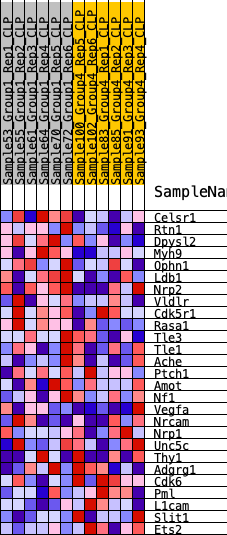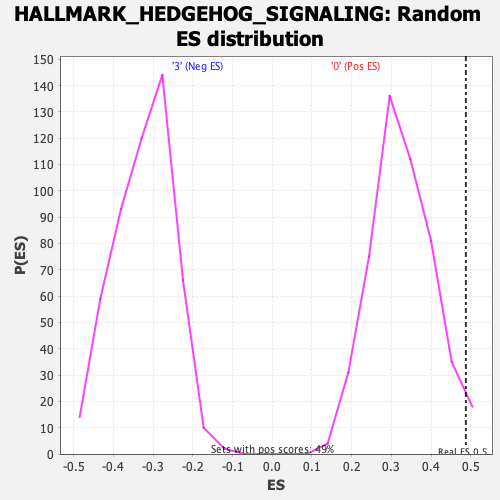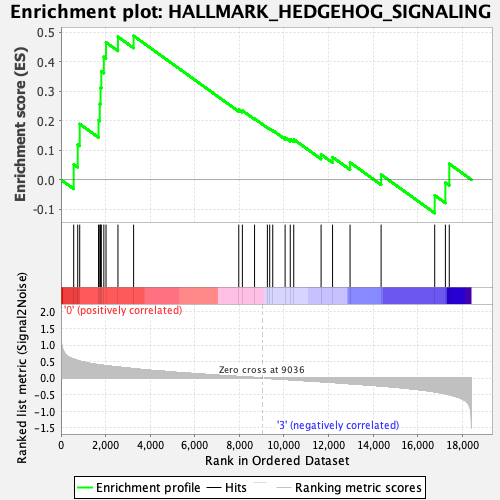
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group1_versus_Group4.Basophil_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_HEDGEHOG_SIGNALING |
| Enrichment Score (ES) | 0.4875886 |
| Normalized Enrichment Score (NES) | 1.4903808 |
| Nominal p-value | 0.026422765 |
| FDR q-value | 0.5324932 |
| FWER p-Value | 0.41 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Celsr1 | 569 | 0.568 | 0.0520 | Yes |
| 2 | Rtn1 | 750 | 0.530 | 0.1195 | Yes |
| 3 | Dpysl2 | 839 | 0.509 | 0.1890 | Yes |
| 4 | Myh9 | 1683 | 0.402 | 0.2018 | Yes |
| 5 | Ophn1 | 1739 | 0.397 | 0.2568 | Yes |
| 6 | Ldb1 | 1773 | 0.392 | 0.3122 | Yes |
| 7 | Nrp2 | 1808 | 0.388 | 0.3670 | Yes |
| 8 | Vldlr | 1921 | 0.381 | 0.4165 | Yes |
| 9 | Cdk5r1 | 2019 | 0.372 | 0.4656 | Yes |
| 10 | Rasa1 | 2554 | 0.328 | 0.4845 | Yes |
| 11 | Tle3 | 3256 | 0.282 | 0.4876 | Yes |
| 12 | Tle1 | 7972 | 0.044 | 0.2375 | No |
| 13 | Ache | 8134 | 0.038 | 0.2343 | No |
| 14 | Ptch1 | 8680 | 0.016 | 0.2071 | No |
| 15 | Amot | 9255 | -0.005 | 0.1766 | No |
| 16 | Nf1 | 9359 | -0.010 | 0.1724 | No |
| 17 | Vegfa | 9496 | -0.016 | 0.1673 | No |
| 18 | Nrcam | 10050 | -0.038 | 0.1427 | No |
| 19 | Nrp1 | 10281 | -0.048 | 0.1373 | No |
| 20 | Unc5c | 10437 | -0.055 | 0.1369 | No |
| 21 | Thy1 | 11664 | -0.107 | 0.0858 | No |
| 22 | Adgrg1 | 12178 | -0.129 | 0.0767 | No |
| 23 | Cdk6 | 12963 | -0.168 | 0.0586 | No |
| 24 | Pml | 14355 | -0.238 | 0.0176 | No |
| 25 | L1cam | 16759 | -0.413 | -0.0527 | No |
| 26 | Slit1 | 17238 | -0.473 | -0.0096 | No |
| 27 | Ets2 | 17412 | -0.502 | 0.0542 | No |