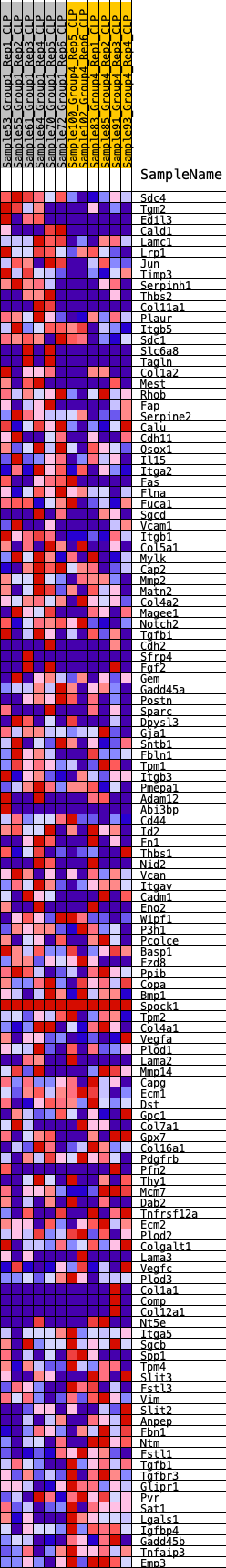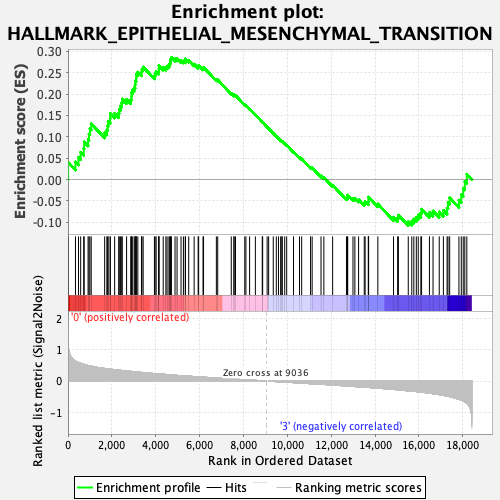
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group1_versus_Group4.Basophil_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION |
| Enrichment Score (ES) | 0.2859148 |
| Normalized Enrichment Score (NES) | 1.1813264 |
| Nominal p-value | 0.19472617 |
| FDR q-value | 0.56723374 |
| FWER p-Value | 0.955 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Sdc4 | 6 | 1.325 | 0.0402 | Yes |
| 2 | Tgm2 | 341 | 0.635 | 0.0414 | Yes |
| 3 | Edil3 | 478 | 0.593 | 0.0521 | Yes |
| 4 | Cald1 | 578 | 0.566 | 0.0640 | Yes |
| 5 | Lamc1 | 717 | 0.535 | 0.0729 | Yes |
| 6 | Lrp1 | 736 | 0.532 | 0.0882 | Yes |
| 7 | Jun | 915 | 0.498 | 0.0937 | Yes |
| 8 | Timp3 | 957 | 0.492 | 0.1065 | Yes |
| 9 | Serpinh1 | 999 | 0.487 | 0.1191 | Yes |
| 10 | Thbs2 | 1058 | 0.478 | 0.1306 | Yes |
| 11 | Col11a1 | 1671 | 0.402 | 0.1094 | Yes |
| 12 | Plaur | 1771 | 0.392 | 0.1160 | Yes |
| 13 | Itgb5 | 1807 | 0.388 | 0.1259 | Yes |
| 14 | Sdc1 | 1831 | 0.386 | 0.1365 | Yes |
| 15 | Slc6a8 | 1923 | 0.381 | 0.1432 | Yes |
| 16 | Tagln | 1927 | 0.381 | 0.1547 | Yes |
| 17 | Col1a2 | 2126 | 0.363 | 0.1550 | Yes |
| 18 | Mest | 2312 | 0.348 | 0.1555 | Yes |
| 19 | Rhob | 2337 | 0.346 | 0.1648 | Yes |
| 20 | Fap | 2397 | 0.342 | 0.1720 | Yes |
| 21 | Serpine2 | 2450 | 0.338 | 0.1795 | Yes |
| 22 | Calu | 2472 | 0.336 | 0.1887 | Yes |
| 23 | Cdh11 | 2671 | 0.324 | 0.1878 | Yes |
| 24 | Qsox1 | 2859 | 0.310 | 0.1870 | Yes |
| 25 | Il15 | 2894 | 0.307 | 0.1946 | Yes |
| 26 | Itga2 | 2899 | 0.306 | 0.2037 | Yes |
| 27 | Fas | 2949 | 0.303 | 0.2103 | Yes |
| 28 | Flna | 3020 | 0.299 | 0.2156 | Yes |
| 29 | Fuca1 | 3061 | 0.295 | 0.2225 | Yes |
| 30 | Sgcd | 3069 | 0.294 | 0.2311 | Yes |
| 31 | Vcam1 | 3110 | 0.291 | 0.2378 | Yes |
| 32 | Itgb1 | 3112 | 0.291 | 0.2467 | Yes |
| 33 | Col5a1 | 3182 | 0.286 | 0.2517 | Yes |
| 34 | Mylk | 3349 | 0.275 | 0.2510 | Yes |
| 35 | Cap2 | 3369 | 0.273 | 0.2583 | Yes |
| 36 | Mmp2 | 3426 | 0.269 | 0.2635 | Yes |
| 37 | Matn2 | 3939 | 0.237 | 0.2427 | Yes |
| 38 | Col4a2 | 3969 | 0.235 | 0.2484 | Yes |
| 39 | Magee1 | 4019 | 0.231 | 0.2528 | Yes |
| 40 | Notch2 | 4135 | 0.224 | 0.2533 | Yes |
| 41 | Tgfbi | 4142 | 0.224 | 0.2598 | Yes |
| 42 | Cdh2 | 4145 | 0.224 | 0.2666 | Yes |
| 43 | Sfrp4 | 4340 | 0.216 | 0.2626 | Yes |
| 44 | Fgf2 | 4462 | 0.211 | 0.2624 | Yes |
| 45 | Gem | 4516 | 0.208 | 0.2659 | Yes |
| 46 | Gadd45a | 4602 | 0.203 | 0.2674 | Yes |
| 47 | Postn | 4638 | 0.201 | 0.2717 | Yes |
| 48 | Sparc | 4660 | 0.200 | 0.2767 | Yes |
| 49 | Dpysl3 | 4679 | 0.199 | 0.2818 | Yes |
| 50 | Gja1 | 4714 | 0.197 | 0.2859 | Yes |
| 51 | Sntb1 | 4879 | 0.187 | 0.2827 | No |
| 52 | Fbln1 | 4971 | 0.181 | 0.2832 | No |
| 53 | Tpm1 | 5153 | 0.171 | 0.2785 | No |
| 54 | Itgb3 | 5261 | 0.165 | 0.2777 | No |
| 55 | Pmepa1 | 5348 | 0.162 | 0.2780 | No |
| 56 | Adam12 | 5351 | 0.161 | 0.2828 | No |
| 57 | Abi3bp | 5503 | 0.154 | 0.2793 | No |
| 58 | Cd44 | 5750 | 0.143 | 0.2702 | No |
| 59 | Id2 | 5933 | 0.135 | 0.2644 | No |
| 60 | Fn1 | 5956 | 0.135 | 0.2673 | No |
| 61 | Thbs1 | 6155 | 0.124 | 0.2603 | No |
| 62 | Nid2 | 6179 | 0.124 | 0.2628 | No |
| 63 | Vcan | 6763 | 0.096 | 0.2339 | No |
| 64 | Itgav | 6825 | 0.093 | 0.2334 | No |
| 65 | Cadm1 | 7442 | 0.067 | 0.2018 | No |
| 66 | Eno2 | 7564 | 0.061 | 0.1970 | No |
| 67 | Wipf1 | 7570 | 0.061 | 0.1986 | No |
| 68 | P3h1 | 7635 | 0.058 | 0.1969 | No |
| 69 | Pcolce | 8058 | 0.041 | 0.1750 | No |
| 70 | Basp1 | 8110 | 0.039 | 0.1735 | No |
| 71 | Fzd8 | 8279 | 0.032 | 0.1652 | No |
| 72 | Ppib | 8541 | 0.022 | 0.1516 | No |
| 73 | Copa | 8855 | 0.009 | 0.1348 | No |
| 74 | Bmp1 | 8874 | 0.008 | 0.1340 | No |
| 75 | Spock1 | 9078 | 0.000 | 0.1229 | No |
| 76 | Tpm2 | 9146 | -0.000 | 0.1193 | No |
| 77 | Col4a1 | 9357 | -0.010 | 0.1081 | No |
| 78 | Vegfa | 9496 | -0.016 | 0.1010 | No |
| 79 | Plod1 | 9594 | -0.019 | 0.0963 | No |
| 80 | Lama2 | 9693 | -0.023 | 0.0916 | No |
| 81 | Mmp14 | 9747 | -0.026 | 0.0895 | No |
| 82 | Capg | 9768 | -0.027 | 0.0892 | No |
| 83 | Ecm1 | 9879 | -0.031 | 0.0842 | No |
| 84 | Dst | 9969 | -0.034 | 0.0803 | No |
| 85 | Gpc1 | 10280 | -0.048 | 0.0649 | No |
| 86 | Col7a1 | 10553 | -0.060 | 0.0518 | No |
| 87 | Gpx7 | 10652 | -0.064 | 0.0484 | No |
| 88 | Col16a1 | 11058 | -0.082 | 0.0287 | No |
| 89 | Pdgfrb | 11130 | -0.084 | 0.0274 | No |
| 90 | Pfn2 | 11534 | -0.102 | 0.0085 | No |
| 91 | Thy1 | 11664 | -0.107 | 0.0047 | No |
| 92 | Mcm7 | 12064 | -0.124 | -0.0133 | No |
| 93 | Dab2 | 12694 | -0.154 | -0.0430 | No |
| 94 | Tnfrsf12a | 12743 | -0.156 | -0.0408 | No |
| 95 | Ecm2 | 12750 | -0.156 | -0.0364 | No |
| 96 | Plod2 | 12991 | -0.169 | -0.0443 | No |
| 97 | Colgalt1 | 13077 | -0.173 | -0.0436 | No |
| 98 | Lama3 | 13246 | -0.182 | -0.0473 | No |
| 99 | Vegfc | 13505 | -0.194 | -0.0554 | No |
| 100 | Plod3 | 13545 | -0.196 | -0.0515 | No |
| 101 | Col1a1 | 13688 | -0.205 | -0.0530 | No |
| 102 | Comp | 13701 | -0.205 | -0.0474 | No |
| 103 | Col12a1 | 13702 | -0.205 | -0.0411 | No |
| 104 | Nt5e | 14124 | -0.225 | -0.0573 | No |
| 105 | Itga5 | 14839 | -0.265 | -0.0882 | No |
| 106 | Sgcb | 15026 | -0.274 | -0.0900 | No |
| 107 | Spp1 | 15060 | -0.276 | -0.0834 | No |
| 108 | Tpm4 | 15511 | -0.308 | -0.0985 | No |
| 109 | Slit3 | 15672 | -0.321 | -0.0975 | No |
| 110 | Fstl3 | 15765 | -0.327 | -0.0925 | No |
| 111 | Vim | 15878 | -0.335 | -0.0884 | No |
| 112 | Slit2 | 15966 | -0.342 | -0.0827 | No |
| 113 | Anpep | 16085 | -0.351 | -0.0784 | No |
| 114 | Fbn1 | 16118 | -0.353 | -0.0693 | No |
| 115 | Ntm | 16476 | -0.386 | -0.0770 | No |
| 116 | Fstl1 | 16640 | -0.401 | -0.0736 | No |
| 117 | Tgfb1 | 16923 | -0.431 | -0.0759 | No |
| 118 | Tgfbr3 | 17112 | -0.458 | -0.0721 | No |
| 119 | Glipr1 | 17281 | -0.478 | -0.0667 | No |
| 120 | Pvr | 17322 | -0.485 | -0.0540 | No |
| 121 | Sat1 | 17399 | -0.498 | -0.0429 | No |
| 122 | Lgals1 | 17821 | -0.581 | -0.0482 | No |
| 123 | Igfbp4 | 17928 | -0.606 | -0.0354 | No |
| 124 | Gadd45b | 18016 | -0.636 | -0.0207 | No |
| 125 | Tnfaip3 | 18088 | -0.664 | -0.0042 | No |
| 126 | Emp3 | 18181 | -0.709 | 0.0124 | No |