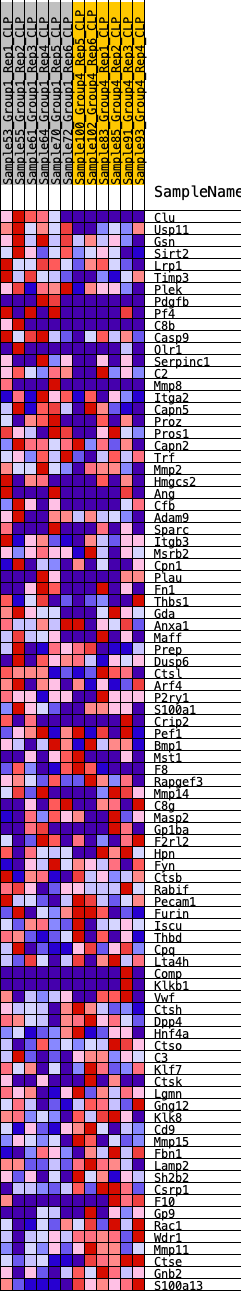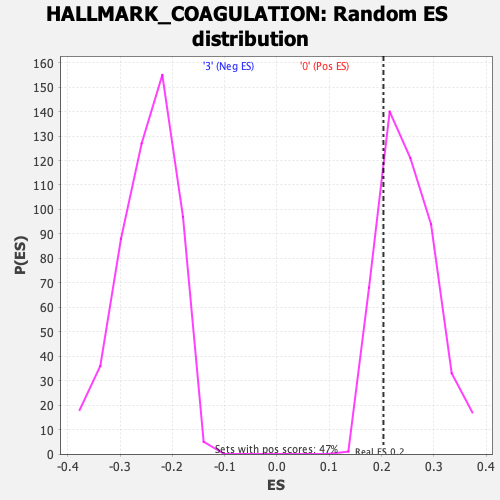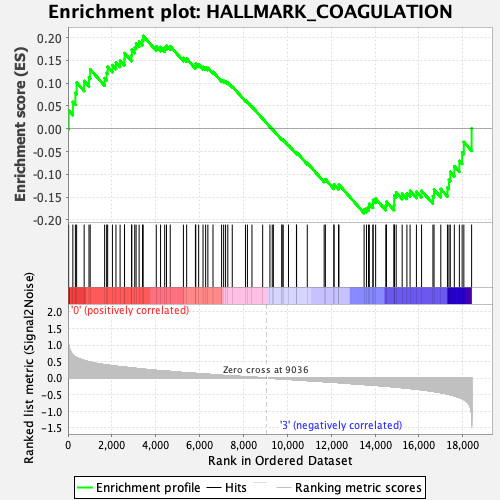
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group1_versus_Group4.Basophil_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_COAGULATION |
| Enrichment Score (ES) | 0.20368375 |
| Normalized Enrichment Score (NES) | 0.8145527 |
| Nominal p-value | 0.8016878 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Clu | 30 | 1.010 | 0.0402 | Yes |
| 2 | Usp11 | 221 | 0.699 | 0.0588 | Yes |
| 3 | Gsn | 337 | 0.636 | 0.0788 | Yes |
| 4 | Sirt2 | 394 | 0.616 | 0.1013 | Yes |
| 5 | Lrp1 | 736 | 0.532 | 0.1047 | Yes |
| 6 | Timp3 | 957 | 0.492 | 0.1131 | Yes |
| 7 | Plek | 1011 | 0.485 | 0.1303 | Yes |
| 8 | Pdgfb | 1670 | 0.402 | 0.1110 | Yes |
| 9 | Pf4 | 1762 | 0.393 | 0.1223 | Yes |
| 10 | C8b | 1809 | 0.388 | 0.1359 | Yes |
| 11 | Casp9 | 2025 | 0.372 | 0.1395 | Yes |
| 12 | Olr1 | 2186 | 0.359 | 0.1457 | Yes |
| 13 | Serpinc1 | 2375 | 0.344 | 0.1496 | Yes |
| 14 | C2 | 2574 | 0.327 | 0.1524 | Yes |
| 15 | Mmp8 | 2577 | 0.327 | 0.1658 | Yes |
| 16 | Itga2 | 2899 | 0.306 | 0.1610 | Yes |
| 17 | Capn5 | 2907 | 0.306 | 0.1733 | Yes |
| 18 | Proz | 3043 | 0.297 | 0.1782 | Yes |
| 19 | Pros1 | 3104 | 0.292 | 0.1870 | Yes |
| 20 | Capn2 | 3242 | 0.283 | 0.1912 | Yes |
| 21 | Trf | 3396 | 0.271 | 0.1941 | Yes |
| 22 | Mmp2 | 3426 | 0.269 | 0.2037 | Yes |
| 23 | Hmgcs2 | 4024 | 0.231 | 0.1807 | No |
| 24 | Ang | 4219 | 0.220 | 0.1792 | No |
| 25 | Cfb | 4408 | 0.214 | 0.1778 | No |
| 26 | Adam9 | 4490 | 0.210 | 0.1821 | No |
| 27 | Sparc | 4660 | 0.200 | 0.1811 | No |
| 28 | Itgb3 | 5261 | 0.165 | 0.1552 | No |
| 29 | Msrb2 | 5409 | 0.158 | 0.1537 | No |
| 30 | Cpn1 | 5808 | 0.141 | 0.1378 | No |
| 31 | Plau | 5824 | 0.140 | 0.1428 | No |
| 32 | Fn1 | 5956 | 0.135 | 0.1412 | No |
| 33 | Thbs1 | 6155 | 0.124 | 0.1356 | No |
| 34 | Gda | 6274 | 0.119 | 0.1341 | No |
| 35 | Anxa1 | 6376 | 0.114 | 0.1333 | No |
| 36 | Maff | 6610 | 0.103 | 0.1248 | No |
| 37 | Prep | 7002 | 0.084 | 0.1070 | No |
| 38 | Dusp6 | 7095 | 0.081 | 0.1053 | No |
| 39 | Ctsl | 7183 | 0.078 | 0.1038 | No |
| 40 | Arf4 | 7285 | 0.073 | 0.1013 | No |
| 41 | P2ry1 | 7488 | 0.064 | 0.0929 | No |
| 42 | S100a1 | 8090 | 0.040 | 0.0618 | No |
| 43 | Crip2 | 8179 | 0.037 | 0.0585 | No |
| 44 | Pef1 | 8386 | 0.028 | 0.0484 | No |
| 45 | Bmp1 | 8874 | 0.008 | 0.0221 | No |
| 46 | Mst1 | 9205 | -0.003 | 0.0043 | No |
| 47 | F8 | 9311 | -0.008 | -0.0012 | No |
| 48 | Rapgef3 | 9362 | -0.010 | -0.0035 | No |
| 49 | Mmp14 | 9747 | -0.026 | -0.0234 | No |
| 50 | C8g | 9756 | -0.026 | -0.0227 | No |
| 51 | Masp2 | 9811 | -0.028 | -0.0245 | No |
| 52 | Gp1ba | 10051 | -0.038 | -0.0360 | No |
| 53 | F2rl2 | 10414 | -0.054 | -0.0535 | No |
| 54 | Hpn | 10421 | -0.054 | -0.0516 | No |
| 55 | Fyn | 10908 | -0.075 | -0.0750 | No |
| 56 | Ctsb | 11677 | -0.107 | -0.1125 | No |
| 57 | Rabif | 11733 | -0.110 | -0.1109 | No |
| 58 | Pecam1 | 12121 | -0.127 | -0.1268 | No |
| 59 | Furin | 12127 | -0.127 | -0.1218 | No |
| 60 | Iscu | 12332 | -0.136 | -0.1273 | No |
| 61 | Thbd | 12351 | -0.138 | -0.1226 | No |
| 62 | Cpq | 13498 | -0.194 | -0.1771 | No |
| 63 | Lta4h | 13606 | -0.200 | -0.1747 | No |
| 64 | Comp | 13701 | -0.205 | -0.1713 | No |
| 65 | Klkb1 | 13735 | -0.205 | -0.1646 | No |
| 66 | Vwf | 13903 | -0.213 | -0.1649 | No |
| 67 | Ctsh | 13905 | -0.213 | -0.1562 | No |
| 68 | Dpp4 | 14024 | -0.220 | -0.1535 | No |
| 69 | Hnf4a | 14488 | -0.245 | -0.1686 | No |
| 70 | Ctso | 14517 | -0.247 | -0.1599 | No |
| 71 | C3 | 14861 | -0.266 | -0.1676 | No |
| 72 | Klf7 | 14873 | -0.267 | -0.1572 | No |
| 73 | Ctsk | 14874 | -0.267 | -0.1461 | No |
| 74 | Lgmn | 14961 | -0.272 | -0.1396 | No |
| 75 | Gng12 | 15233 | -0.291 | -0.1423 | No |
| 76 | Klk8 | 15449 | -0.306 | -0.1414 | No |
| 77 | Cd9 | 15594 | -0.315 | -0.1362 | No |
| 78 | Mmp15 | 15883 | -0.335 | -0.1381 | No |
| 79 | Fbn1 | 16118 | -0.353 | -0.1362 | No |
| 80 | Lamp2 | 16636 | -0.401 | -0.1478 | No |
| 81 | Sh2b2 | 16686 | -0.407 | -0.1336 | No |
| 82 | Csrp1 | 16994 | -0.443 | -0.1321 | No |
| 83 | F10 | 17298 | -0.480 | -0.1287 | No |
| 84 | Gp9 | 17366 | -0.492 | -0.1120 | No |
| 85 | Rac1 | 17427 | -0.503 | -0.0945 | No |
| 86 | Wdr1 | 17611 | -0.536 | -0.0823 | No |
| 87 | Mmp11 | 17841 | -0.586 | -0.0705 | No |
| 88 | Ctse | 17967 | -0.620 | -0.0516 | No |
| 89 | Gnb2 | 18045 | -0.648 | -0.0290 | No |
| 90 | S100a13 | 18399 | -1.177 | 0.0005 | No |