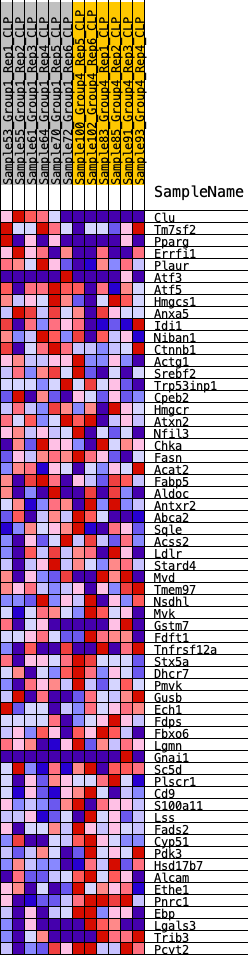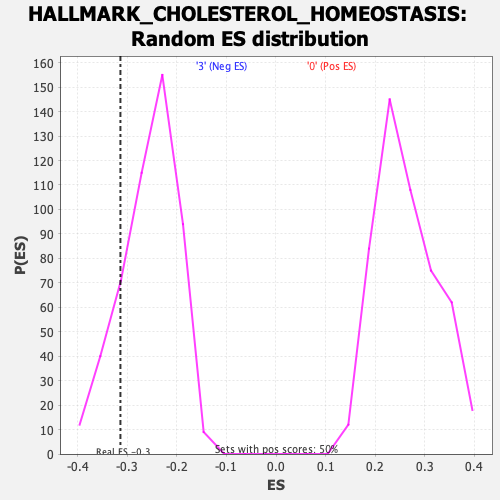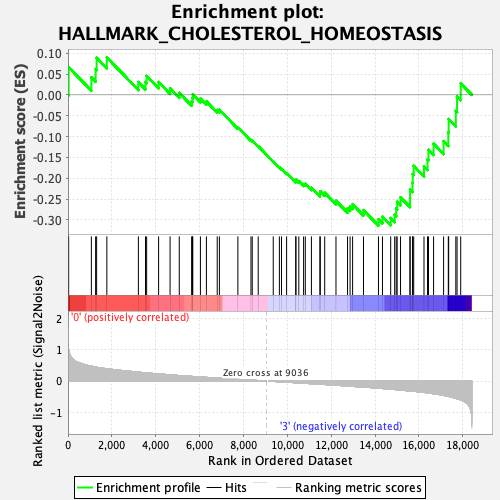
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group1_versus_Group4.Basophil_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_CHOLESTEROL_HOMEOSTASIS |
| Enrichment Score (ES) | -0.31356767 |
| Normalized Enrichment Score (NES) | -1.2226588 |
| Nominal p-value | 0.16129032 |
| FDR q-value | 0.41365787 |
| FWER p-Value | 0.921 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Clu | 30 | 1.010 | 0.0663 | No |
| 2 | Tm7sf2 | 1065 | 0.477 | 0.0421 | No |
| 3 | Pparg | 1259 | 0.447 | 0.0616 | No |
| 4 | Errfi1 | 1305 | 0.440 | 0.0888 | No |
| 5 | Plaur | 1771 | 0.392 | 0.0898 | No |
| 6 | Atf3 | 3208 | 0.285 | 0.0307 | No |
| 7 | Atf5 | 3533 | 0.262 | 0.0306 | No |
| 8 | Hmgcs1 | 3585 | 0.259 | 0.0453 | No |
| 9 | Anxa5 | 4130 | 0.225 | 0.0307 | No |
| 10 | Idi1 | 4653 | 0.200 | 0.0158 | No |
| 11 | Niban1 | 5072 | 0.175 | 0.0048 | No |
| 12 | Ctnnb1 | 5635 | 0.150 | -0.0158 | No |
| 13 | Actg1 | 5675 | 0.147 | -0.0080 | No |
| 14 | Srebf2 | 5688 | 0.146 | 0.0012 | No |
| 15 | Trp53inp1 | 6036 | 0.131 | -0.0089 | No |
| 16 | Cpeb2 | 6309 | 0.117 | -0.0158 | No |
| 17 | Hmgcr | 6802 | 0.095 | -0.0363 | No |
| 18 | Atxn2 | 6901 | 0.090 | -0.0356 | No |
| 19 | Nfil3 | 7744 | 0.054 | -0.0778 | No |
| 20 | Chka | 8341 | 0.030 | -0.1083 | No |
| 21 | Fasn | 8399 | 0.027 | -0.1096 | No |
| 22 | Acat2 | 8671 | 0.017 | -0.1232 | No |
| 23 | Fabp5 | 9356 | -0.009 | -0.1599 | No |
| 24 | Aldoc | 9636 | -0.021 | -0.1737 | No |
| 25 | Antxr2 | 9732 | -0.025 | -0.1772 | No |
| 26 | Abca2 | 9965 | -0.034 | -0.1875 | No |
| 27 | Sqle | 10380 | -0.053 | -0.2065 | No |
| 28 | Acss2 | 10392 | -0.053 | -0.2036 | No |
| 29 | Ldlr | 10522 | -0.059 | -0.2066 | No |
| 30 | Stard4 | 10740 | -0.068 | -0.2139 | No |
| 31 | Mvd | 10818 | -0.071 | -0.2133 | No |
| 32 | Tmem97 | 11096 | -0.083 | -0.2228 | No |
| 33 | Nsdhl | 11483 | -0.100 | -0.2371 | No |
| 34 | Mvk | 11500 | -0.102 | -0.2312 | No |
| 35 | Gstm7 | 11702 | -0.109 | -0.2348 | No |
| 36 | Fdft1 | 12216 | -0.130 | -0.2540 | No |
| 37 | Tnfrsf12a | 12743 | -0.156 | -0.2722 | No |
| 38 | Stx5a | 12859 | -0.163 | -0.2675 | No |
| 39 | Dhcr7 | 12974 | -0.169 | -0.2624 | No |
| 40 | Pmvk | 13470 | -0.192 | -0.2764 | No |
| 41 | Gusb | 14153 | -0.226 | -0.2984 | Yes |
| 42 | Ech1 | 14335 | -0.237 | -0.2923 | Yes |
| 43 | Fdps | 14710 | -0.257 | -0.2954 | Yes |
| 44 | Fbxo6 | 14892 | -0.269 | -0.2872 | Yes |
| 45 | Lgmn | 14961 | -0.272 | -0.2726 | Yes |
| 46 | Gnai1 | 15004 | -0.273 | -0.2565 | Yes |
| 47 | Sc5d | 15159 | -0.284 | -0.2458 | Yes |
| 48 | Plscr1 | 15590 | -0.314 | -0.2481 | Yes |
| 49 | Cd9 | 15594 | -0.315 | -0.2271 | Yes |
| 50 | S100a11 | 15704 | -0.323 | -0.2113 | Yes |
| 51 | Lss | 15713 | -0.324 | -0.1899 | Yes |
| 52 | Fads2 | 15759 | -0.327 | -0.1704 | Yes |
| 53 | Cyp51 | 16228 | -0.361 | -0.1716 | Yes |
| 54 | Pdk3 | 16389 | -0.376 | -0.1550 | Yes |
| 55 | Hsd17b7 | 16433 | -0.381 | -0.1317 | Yes |
| 56 | Alcam | 16665 | -0.403 | -0.1172 | Yes |
| 57 | Ethe1 | 17122 | -0.459 | -0.1111 | Yes |
| 58 | Pnrc1 | 17336 | -0.486 | -0.0900 | Yes |
| 59 | Ebp | 17353 | -0.490 | -0.0579 | Yes |
| 60 | Lgals3 | 17679 | -0.551 | -0.0386 | Yes |
| 61 | Trib3 | 17735 | -0.563 | -0.0037 | Yes |
| 62 | Pcyt2 | 17902 | -0.599 | 0.0276 | Yes |