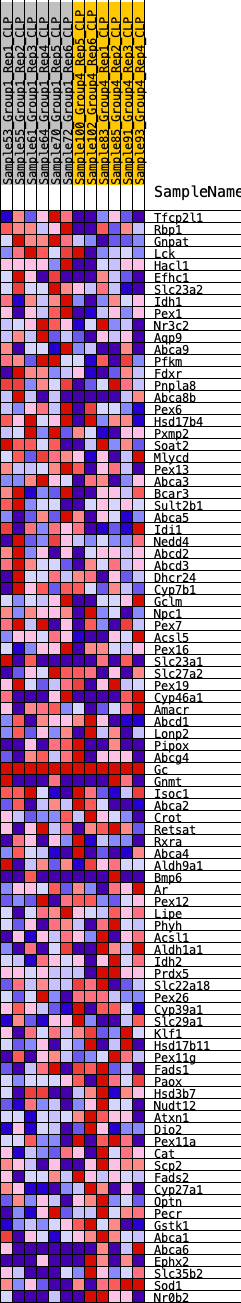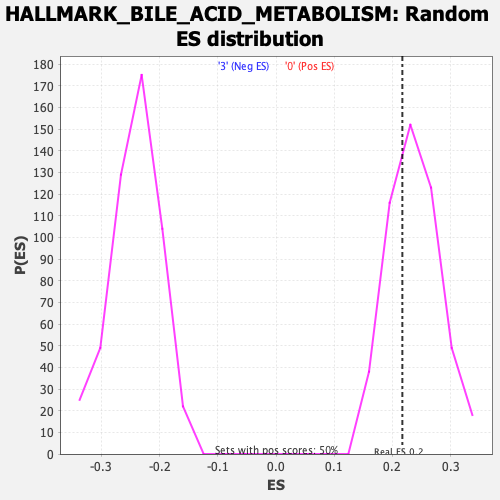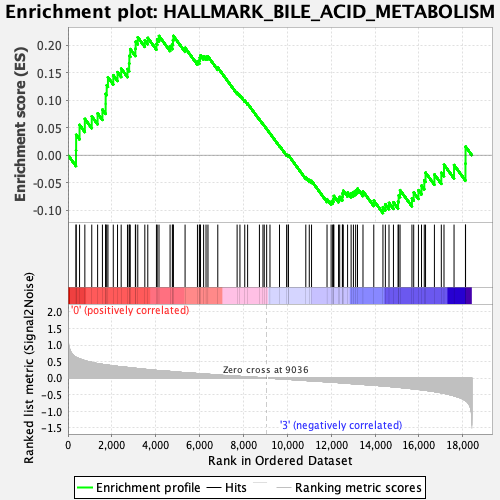
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group1_versus_Group4.Basophil_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_BILE_ACID_METABOLISM |
| Enrichment Score (ES) | 0.21693514 |
| Normalized Enrichment Score (NES) | 0.91944396 |
| Nominal p-value | 0.6532258 |
| FDR q-value | 0.89518726 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Tfcp2l1 | 365 | 0.627 | 0.0088 | Yes |
| 2 | Rbp1 | 371 | 0.624 | 0.0371 | Yes |
| 3 | Gnpat | 527 | 0.578 | 0.0551 | Yes |
| 4 | Lck | 768 | 0.525 | 0.0660 | Yes |
| 5 | Hacl1 | 1083 | 0.473 | 0.0706 | Yes |
| 6 | Efhc1 | 1350 | 0.435 | 0.0759 | Yes |
| 7 | Slc23a2 | 1569 | 0.414 | 0.0830 | Yes |
| 8 | Idh1 | 1716 | 0.399 | 0.0933 | Yes |
| 9 | Pex1 | 1717 | 0.399 | 0.1115 | Yes |
| 10 | Nr3c2 | 1765 | 0.393 | 0.1270 | Yes |
| 11 | Aqp9 | 1821 | 0.387 | 0.1417 | Yes |
| 12 | Abca9 | 2064 | 0.370 | 0.1454 | Yes |
| 13 | Pfkm | 2258 | 0.352 | 0.1510 | Yes |
| 14 | Fdxr | 2424 | 0.340 | 0.1575 | Yes |
| 15 | Pnpla8 | 2711 | 0.321 | 0.1566 | Yes |
| 16 | Abca8b | 2787 | 0.315 | 0.1669 | Yes |
| 17 | Pex6 | 2797 | 0.315 | 0.1808 | Yes |
| 18 | Hsd17b4 | 2838 | 0.312 | 0.1929 | Yes |
| 19 | Pxmp2 | 3071 | 0.294 | 0.1937 | Yes |
| 20 | Soat2 | 3085 | 0.293 | 0.2064 | Yes |
| 21 | Mlycd | 3181 | 0.286 | 0.2143 | Yes |
| 22 | Pex13 | 3502 | 0.263 | 0.2089 | Yes |
| 23 | Abca3 | 3637 | 0.255 | 0.2133 | Yes |
| 24 | Bcar3 | 4031 | 0.231 | 0.2024 | Yes |
| 25 | Sult2b1 | 4066 | 0.229 | 0.2110 | Yes |
| 26 | Abca5 | 4148 | 0.223 | 0.2168 | Yes |
| 27 | Idi1 | 4653 | 0.200 | 0.1984 | Yes |
| 28 | Nedd4 | 4758 | 0.194 | 0.2017 | Yes |
| 29 | Abcd2 | 4785 | 0.193 | 0.2091 | Yes |
| 30 | Abcd3 | 4803 | 0.192 | 0.2169 | Yes |
| 31 | Dhcr24 | 5337 | 0.162 | 0.1952 | No |
| 32 | Cyp7b1 | 5901 | 0.137 | 0.1708 | No |
| 33 | Gclm | 5987 | 0.134 | 0.1723 | No |
| 34 | Npc1 | 6010 | 0.133 | 0.1771 | No |
| 35 | Pex7 | 6040 | 0.131 | 0.1815 | No |
| 36 | Acsl5 | 6182 | 0.124 | 0.1795 | No |
| 37 | Pex16 | 6292 | 0.118 | 0.1790 | No |
| 38 | Slc23a1 | 6380 | 0.114 | 0.1794 | No |
| 39 | Slc27a2 | 6828 | 0.093 | 0.1593 | No |
| 40 | Pex19 | 7709 | 0.055 | 0.1138 | No |
| 41 | Cyp46a1 | 7838 | 0.050 | 0.1091 | No |
| 42 | Amacr | 8057 | 0.041 | 0.0991 | No |
| 43 | Abcd1 | 8188 | 0.036 | 0.0936 | No |
| 44 | Lonp2 | 8726 | 0.014 | 0.0650 | No |
| 45 | Pipox | 8887 | 0.007 | 0.0565 | No |
| 46 | Abcg4 | 8946 | 0.005 | 0.0536 | No |
| 47 | Gc | 9056 | 0.000 | 0.0476 | No |
| 48 | Gnmt | 9207 | -0.003 | 0.0396 | No |
| 49 | Isoc1 | 9641 | -0.021 | 0.0169 | No |
| 50 | Abca2 | 9965 | -0.034 | 0.0008 | No |
| 51 | Crot | 10038 | -0.037 | -0.0014 | No |
| 52 | Retsat | 10047 | -0.038 | -0.0001 | No |
| 53 | Rxra | 10840 | -0.072 | -0.0400 | No |
| 54 | Abca4 | 10999 | -0.079 | -0.0450 | No |
| 55 | Aldh9a1 | 11102 | -0.083 | -0.0468 | No |
| 56 | Bmp6 | 11808 | -0.113 | -0.0801 | No |
| 57 | Ar | 11994 | -0.121 | -0.0847 | No |
| 58 | Pex12 | 12063 | -0.124 | -0.0827 | No |
| 59 | Lipe | 12103 | -0.126 | -0.0790 | No |
| 60 | Phyh | 12120 | -0.127 | -0.0741 | No |
| 61 | Acsl1 | 12333 | -0.137 | -0.0794 | No |
| 62 | Aldh1a1 | 12385 | -0.140 | -0.0758 | No |
| 63 | Idh2 | 12507 | -0.146 | -0.0758 | No |
| 64 | Prdx5 | 12514 | -0.146 | -0.0694 | No |
| 65 | Slc22a18 | 12548 | -0.148 | -0.0645 | No |
| 66 | Pex26 | 12745 | -0.156 | -0.0680 | No |
| 67 | Cyp39a1 | 12906 | -0.166 | -0.0692 | No |
| 68 | Slc29a1 | 13011 | -0.170 | -0.0671 | No |
| 69 | Klf1 | 13113 | -0.175 | -0.0646 | No |
| 70 | Hsd17b11 | 13197 | -0.180 | -0.0609 | No |
| 71 | Pex11g | 13447 | -0.191 | -0.0657 | No |
| 72 | Fads1 | 13939 | -0.215 | -0.0827 | No |
| 73 | Paox | 14357 | -0.238 | -0.0945 | No |
| 74 | Hsd3b7 | 14467 | -0.244 | -0.0893 | No |
| 75 | Nudt12 | 14633 | -0.254 | -0.0867 | No |
| 76 | Atxn1 | 14836 | -0.265 | -0.0856 | No |
| 77 | Dio2 | 15046 | -0.275 | -0.0844 | No |
| 78 | Pex11a | 15074 | -0.278 | -0.0732 | No |
| 79 | Cat | 15137 | -0.283 | -0.0637 | No |
| 80 | Scp2 | 15679 | -0.321 | -0.0785 | No |
| 81 | Fads2 | 15759 | -0.327 | -0.0679 | No |
| 82 | Cyp27a1 | 15975 | -0.343 | -0.0639 | No |
| 83 | Optn | 16115 | -0.353 | -0.0553 | No |
| 84 | Pecr | 16241 | -0.362 | -0.0456 | No |
| 85 | Gstk1 | 16299 | -0.369 | -0.0318 | No |
| 86 | Abca1 | 16703 | -0.408 | -0.0351 | No |
| 87 | Abca6 | 17018 | -0.446 | -0.0318 | No |
| 88 | Ephx2 | 17140 | -0.460 | -0.0174 | No |
| 89 | Slc35b2 | 17598 | -0.533 | -0.0179 | No |
| 90 | Sod1 | 18118 | -0.676 | -0.0153 | No |
| 91 | Nr0b2 | 18123 | -0.678 | 0.0156 | No |