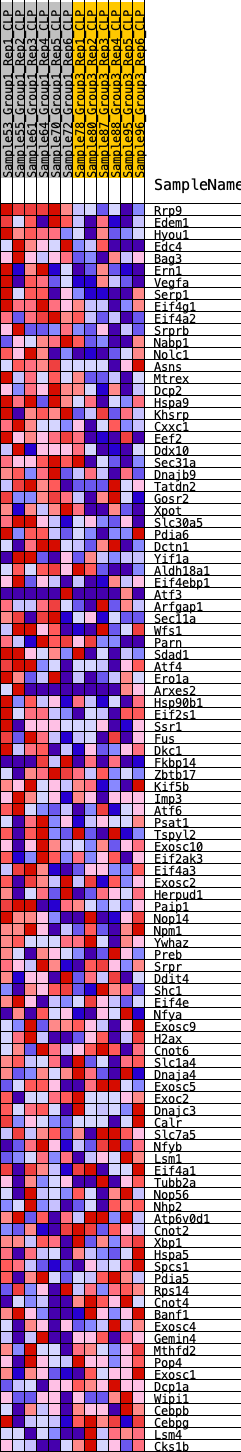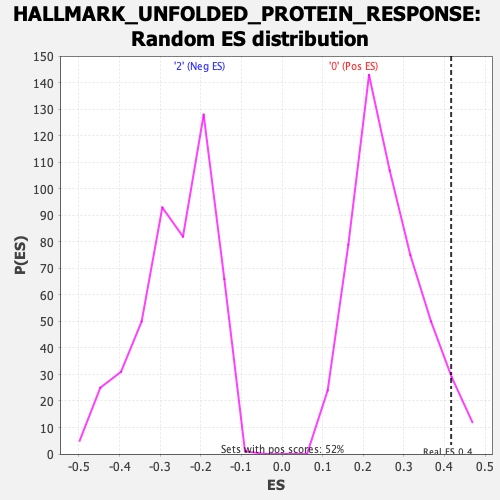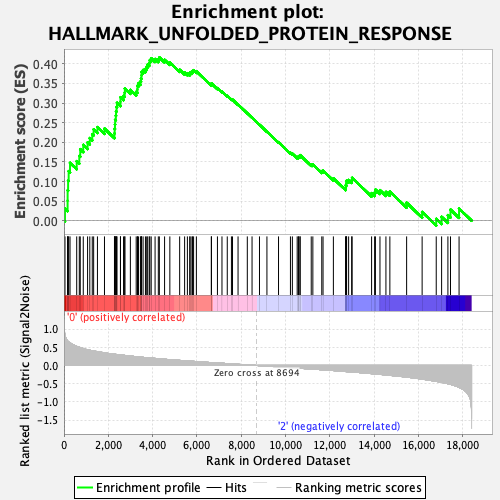
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group1_versus_Group3.Basophil_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_UNFOLDED_PROTEIN_RESPONSE |
| Enrichment Score (ES) | 0.41643155 |
| Normalized Enrichment Score (NES) | 1.6075624 |
| Nominal p-value | 0.05202312 |
| FDR q-value | 0.22768338 |
| FWER p-Value | 0.223 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Rrp9 | 44 | 0.839 | 0.0307 | Yes |
| 2 | Edem1 | 162 | 0.674 | 0.0510 | Yes |
| 3 | Hyou1 | 164 | 0.673 | 0.0775 | Yes |
| 4 | Edc4 | 186 | 0.656 | 0.1022 | Yes |
| 5 | Bag3 | 211 | 0.643 | 0.1263 | Yes |
| 6 | Ern1 | 272 | 0.622 | 0.1476 | Yes |
| 7 | Vegfa | 574 | 0.524 | 0.1518 | Yes |
| 8 | Serp1 | 691 | 0.500 | 0.1652 | Yes |
| 9 | Eif4g1 | 734 | 0.492 | 0.1823 | Yes |
| 10 | Eif4a2 | 870 | 0.466 | 0.1934 | Yes |
| 11 | Srprb | 1065 | 0.429 | 0.1997 | Yes |
| 12 | Nabp1 | 1165 | 0.416 | 0.2107 | Yes |
| 13 | Nolc1 | 1276 | 0.405 | 0.2207 | Yes |
| 14 | Asns | 1339 | 0.397 | 0.2330 | Yes |
| 15 | Mtrex | 1510 | 0.379 | 0.2386 | Yes |
| 16 | Dcp2 | 1828 | 0.346 | 0.2350 | Yes |
| 17 | Hspa9 | 2279 | 0.309 | 0.2226 | Yes |
| 18 | Khsrp | 2287 | 0.308 | 0.2344 | Yes |
| 19 | Cxxc1 | 2302 | 0.307 | 0.2457 | Yes |
| 20 | Eef2 | 2310 | 0.306 | 0.2574 | Yes |
| 21 | Ddx10 | 2341 | 0.303 | 0.2677 | Yes |
| 22 | Sec31a | 2351 | 0.302 | 0.2792 | Yes |
| 23 | Dnajb9 | 2373 | 0.301 | 0.2899 | Yes |
| 24 | Tatdn2 | 2393 | 0.299 | 0.3007 | Yes |
| 25 | Gosr2 | 2549 | 0.287 | 0.3036 | Yes |
| 26 | Xpot | 2551 | 0.287 | 0.3148 | Yes |
| 27 | Slc30a5 | 2689 | 0.279 | 0.3184 | Yes |
| 28 | Pdia6 | 2741 | 0.275 | 0.3264 | Yes |
| 29 | Dctn1 | 2749 | 0.275 | 0.3369 | Yes |
| 30 | Yif1a | 2997 | 0.257 | 0.3335 | Yes |
| 31 | Aldh18a1 | 3260 | 0.241 | 0.3287 | Yes |
| 32 | Eif4ebp1 | 3316 | 0.236 | 0.3351 | Yes |
| 33 | Atf3 | 3318 | 0.236 | 0.3443 | Yes |
| 34 | Arfgap1 | 3366 | 0.232 | 0.3509 | Yes |
| 35 | Sec11a | 3452 | 0.228 | 0.3553 | Yes |
| 36 | Wfs1 | 3478 | 0.227 | 0.3629 | Yes |
| 37 | Parn | 3500 | 0.225 | 0.3706 | Yes |
| 38 | Sdad1 | 3503 | 0.225 | 0.3794 | Yes |
| 39 | Atf4 | 3572 | 0.221 | 0.3844 | Yes |
| 40 | Ero1a | 3678 | 0.216 | 0.3872 | Yes |
| 41 | Arxes2 | 3736 | 0.213 | 0.3925 | Yes |
| 42 | Hsp90b1 | 3779 | 0.211 | 0.3985 | Yes |
| 43 | Eif2s1 | 3851 | 0.207 | 0.4028 | Yes |
| 44 | Ssr1 | 3877 | 0.205 | 0.4095 | Yes |
| 45 | Fus | 3938 | 0.202 | 0.4142 | Yes |
| 46 | Dkc1 | 4113 | 0.193 | 0.4124 | Yes |
| 47 | Fkbp14 | 4259 | 0.185 | 0.4117 | Yes |
| 48 | Zbtb17 | 4306 | 0.182 | 0.4164 | Yes |
| 49 | Kif5b | 4542 | 0.170 | 0.4103 | No |
| 50 | Imp3 | 4780 | 0.158 | 0.4036 | No |
| 51 | Atf6 | 5223 | 0.139 | 0.3849 | No |
| 52 | Psat1 | 5447 | 0.129 | 0.3779 | No |
| 53 | Tspyl2 | 5572 | 0.124 | 0.3760 | No |
| 54 | Exosc10 | 5678 | 0.119 | 0.3750 | No |
| 55 | Eif2ak3 | 5696 | 0.119 | 0.3787 | No |
| 56 | Eif4a3 | 5778 | 0.115 | 0.3788 | No |
| 57 | Exosc2 | 5805 | 0.113 | 0.3819 | No |
| 58 | Herpud1 | 5852 | 0.111 | 0.3838 | No |
| 59 | Paip1 | 5978 | 0.106 | 0.3811 | No |
| 60 | Nop14 | 6654 | 0.078 | 0.3473 | No |
| 61 | Npm1 | 6658 | 0.078 | 0.3502 | No |
| 62 | Ywhaz | 6929 | 0.068 | 0.3382 | No |
| 63 | Preb | 7134 | 0.060 | 0.3294 | No |
| 64 | Srpr | 7373 | 0.050 | 0.3184 | No |
| 65 | Ddit4 | 7561 | 0.042 | 0.3098 | No |
| 66 | Shc1 | 7610 | 0.041 | 0.3088 | No |
| 67 | Eif4e | 7860 | 0.031 | 0.2964 | No |
| 68 | Nfya | 8278 | 0.015 | 0.2743 | No |
| 69 | Exosc9 | 8492 | 0.007 | 0.2629 | No |
| 70 | H2ax | 8828 | -0.006 | 0.2448 | No |
| 71 | Cnot6 | 9164 | -0.019 | 0.2273 | No |
| 72 | Slc1a4 | 9688 | -0.039 | 0.2002 | No |
| 73 | Dnaja4 | 10226 | -0.060 | 0.1733 | No |
| 74 | Exosc5 | 10313 | -0.063 | 0.1711 | No |
| 75 | Exoc2 | 10530 | -0.072 | 0.1621 | No |
| 76 | Dnajc3 | 10583 | -0.074 | 0.1622 | No |
| 77 | Calr | 10596 | -0.075 | 0.1645 | No |
| 78 | Slc7a5 | 10657 | -0.077 | 0.1643 | No |
| 79 | Nfyb | 10663 | -0.078 | 0.1671 | No |
| 80 | Lsm1 | 11165 | -0.098 | 0.1436 | No |
| 81 | Eif4a1 | 11238 | -0.101 | 0.1436 | No |
| 82 | Tubb2a | 11641 | -0.117 | 0.1263 | No |
| 83 | Nop56 | 11698 | -0.119 | 0.1279 | No |
| 84 | Nhp2 | 12165 | -0.139 | 0.1080 | No |
| 85 | Atp6v0d1 | 12717 | -0.166 | 0.0844 | No |
| 86 | Cnot2 | 12725 | -0.167 | 0.0906 | No |
| 87 | Xbp1 | 12756 | -0.168 | 0.0956 | No |
| 88 | Hspa5 | 12762 | -0.168 | 0.1020 | No |
| 89 | Spcs1 | 12854 | -0.172 | 0.1038 | No |
| 90 | Pdia5 | 12991 | -0.178 | 0.1034 | No |
| 91 | Rps14 | 13011 | -0.179 | 0.1094 | No |
| 92 | Cnot4 | 13891 | -0.224 | 0.0703 | No |
| 93 | Banf1 | 14028 | -0.232 | 0.0720 | No |
| 94 | Exosc4 | 14063 | -0.234 | 0.0794 | No |
| 95 | Gemin4 | 14269 | -0.244 | 0.0778 | No |
| 96 | Mthfd2 | 14538 | -0.260 | 0.0734 | No |
| 97 | Pop4 | 14716 | -0.270 | 0.0744 | No |
| 98 | Exosc1 | 15473 | -0.320 | 0.0457 | No |
| 99 | Dcp1a | 16173 | -0.373 | 0.0223 | No |
| 100 | Wipi1 | 16810 | -0.435 | 0.0047 | No |
| 101 | Cebpb | 17053 | -0.464 | 0.0098 | No |
| 102 | Cebpg | 17337 | -0.502 | 0.0142 | No |
| 103 | Lsm4 | 17454 | -0.518 | 0.0283 | No |
| 104 | Cks1b | 17841 | -0.602 | 0.0310 | No |