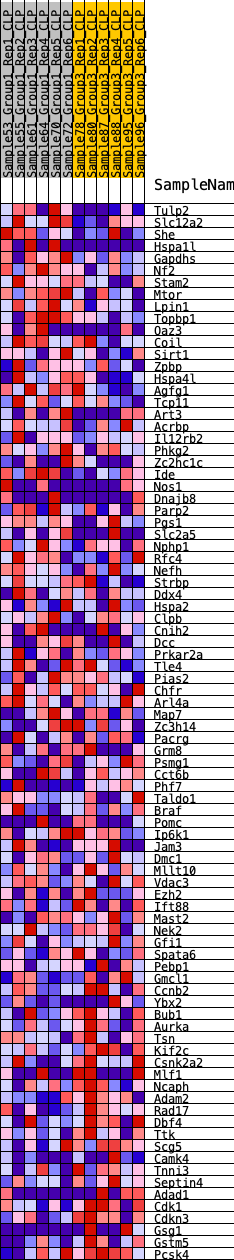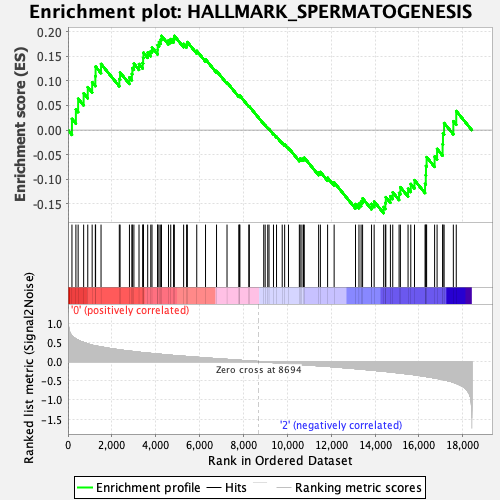
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group1_versus_Group3.Basophil_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_SPERMATOGENESIS |
| Enrichment Score (ES) | 0.19146626 |
| Normalized Enrichment Score (NES) | 0.76469517 |
| Nominal p-value | 0.84732825 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Tulp2 | 180 | 0.659 | 0.0228 | Yes |
| 2 | Slc12a2 | 360 | 0.584 | 0.0419 | Yes |
| 3 | She | 462 | 0.553 | 0.0638 | Yes |
| 4 | Hspa1l | 713 | 0.495 | 0.0746 | Yes |
| 5 | Gapdhs | 901 | 0.460 | 0.0872 | Yes |
| 6 | Nf2 | 1101 | 0.424 | 0.0973 | Yes |
| 7 | Stam2 | 1246 | 0.407 | 0.1096 | Yes |
| 8 | Mtor | 1258 | 0.406 | 0.1291 | Yes |
| 9 | Lpin1 | 1506 | 0.380 | 0.1344 | Yes |
| 10 | Topbp1 | 2339 | 0.303 | 0.1040 | Yes |
| 11 | Oaz3 | 2374 | 0.301 | 0.1170 | Yes |
| 12 | Coil | 2801 | 0.271 | 0.1072 | Yes |
| 13 | Sirt1 | 2913 | 0.263 | 0.1141 | Yes |
| 14 | Zpbp | 2933 | 0.262 | 0.1261 | Yes |
| 15 | Hspa4l | 3004 | 0.256 | 0.1349 | Yes |
| 16 | Agfg1 | 3237 | 0.242 | 0.1342 | Yes |
| 17 | Tcp11 | 3406 | 0.230 | 0.1364 | Yes |
| 18 | Art3 | 3425 | 0.229 | 0.1468 | Yes |
| 19 | Acrbp | 3440 | 0.229 | 0.1573 | Yes |
| 20 | Il12rb2 | 3633 | 0.218 | 0.1576 | Yes |
| 21 | Phkg2 | 3774 | 0.211 | 0.1604 | Yes |
| 22 | Zc2hc1c | 3829 | 0.207 | 0.1677 | Yes |
| 23 | Ide | 4087 | 0.194 | 0.1633 | Yes |
| 24 | Nos1 | 4090 | 0.194 | 0.1728 | Yes |
| 25 | Dnajb8 | 4150 | 0.191 | 0.1790 | Yes |
| 26 | Parp2 | 4227 | 0.187 | 0.1841 | Yes |
| 27 | Pgs1 | 4261 | 0.185 | 0.1915 | Yes |
| 28 | Slc2a5 | 4581 | 0.168 | 0.1824 | No |
| 29 | Nphp1 | 4683 | 0.163 | 0.1849 | No |
| 30 | Rfc4 | 4812 | 0.157 | 0.1857 | No |
| 31 | Nefh | 4848 | 0.154 | 0.1914 | No |
| 32 | Strbp | 5271 | 0.136 | 0.1751 | No |
| 33 | Ddx4 | 5406 | 0.131 | 0.1743 | No |
| 34 | Hspa2 | 5441 | 0.129 | 0.1788 | No |
| 35 | Clpb | 5869 | 0.110 | 0.1610 | No |
| 36 | Cnih2 | 6269 | 0.094 | 0.1438 | No |
| 37 | Dcc | 6771 | 0.074 | 0.1201 | No |
| 38 | Prkar2a | 7249 | 0.055 | 0.0968 | No |
| 39 | Tle4 | 7785 | 0.034 | 0.0693 | No |
| 40 | Pias2 | 7825 | 0.032 | 0.0688 | No |
| 41 | Chfr | 7831 | 0.032 | 0.0701 | No |
| 42 | Arl4a | 8243 | 0.017 | 0.0485 | No |
| 43 | Map7 | 8265 | 0.016 | 0.0481 | No |
| 44 | Zc3h14 | 8923 | -0.010 | 0.0127 | No |
| 45 | Pacrg | 8988 | -0.012 | 0.0098 | No |
| 46 | Grm8 | 9106 | -0.017 | 0.0043 | No |
| 47 | Psmg1 | 9163 | -0.019 | 0.0022 | No |
| 48 | Cct6b | 9367 | -0.026 | -0.0076 | No |
| 49 | Phf7 | 9506 | -0.032 | -0.0136 | No |
| 50 | Taldo1 | 9765 | -0.041 | -0.0256 | No |
| 51 | Braf | 9876 | -0.046 | -0.0294 | No |
| 52 | Pomc | 10051 | -0.052 | -0.0363 | No |
| 53 | Ip6k1 | 10546 | -0.073 | -0.0596 | No |
| 54 | Jam3 | 10582 | -0.074 | -0.0579 | No |
| 55 | Dmc1 | 10664 | -0.078 | -0.0584 | No |
| 56 | Mllt10 | 10735 | -0.081 | -0.0582 | No |
| 57 | Vdac3 | 10770 | -0.082 | -0.0560 | No |
| 58 | Ezh2 | 11425 | -0.108 | -0.0864 | No |
| 59 | Ift88 | 11505 | -0.111 | -0.0852 | No |
| 60 | Mast2 | 11834 | -0.125 | -0.0969 | No |
| 61 | Nek2 | 12132 | -0.137 | -0.1063 | No |
| 62 | Gfi1 | 13106 | -0.183 | -0.1504 | No |
| 63 | Spata6 | 13264 | -0.191 | -0.1495 | No |
| 64 | Pebp1 | 13361 | -0.196 | -0.1451 | No |
| 65 | Gmcl1 | 13429 | -0.200 | -0.1388 | No |
| 66 | Ccnb2 | 13837 | -0.221 | -0.1501 | No |
| 67 | Ybx2 | 13956 | -0.228 | -0.1452 | No |
| 68 | Bub1 | 14389 | -0.251 | -0.1564 | No |
| 69 | Aurka | 14465 | -0.255 | -0.1479 | No |
| 70 | Tsn | 14488 | -0.257 | -0.1364 | No |
| 71 | Kif2c | 14697 | -0.269 | -0.1344 | No |
| 72 | Csnk2a2 | 14804 | -0.276 | -0.1265 | No |
| 73 | Mlf1 | 15093 | -0.295 | -0.1277 | No |
| 74 | Ncaph | 15152 | -0.297 | -0.1161 | No |
| 75 | Adam2 | 15502 | -0.323 | -0.1192 | No |
| 76 | Rad17 | 15626 | -0.330 | -0.1096 | No |
| 77 | Dbf4 | 15795 | -0.343 | -0.1018 | No |
| 78 | Ttk | 16277 | -0.384 | -0.1090 | No |
| 79 | Scg5 | 16310 | -0.387 | -0.0916 | No |
| 80 | Camk4 | 16319 | -0.388 | -0.0729 | No |
| 81 | Tnni3 | 16348 | -0.392 | -0.0550 | No |
| 82 | Septin4 | 16712 | -0.426 | -0.0538 | No |
| 83 | Adad1 | 16825 | -0.437 | -0.0383 | No |
| 84 | Cdk1 | 17077 | -0.467 | -0.0289 | No |
| 85 | Cdkn3 | 17094 | -0.469 | -0.0066 | No |
| 86 | Gsg1 | 17145 | -0.475 | 0.0142 | No |
| 87 | Gstm5 | 17566 | -0.539 | 0.0180 | No |
| 88 | Pcsk4 | 17699 | -0.564 | 0.0387 | No |