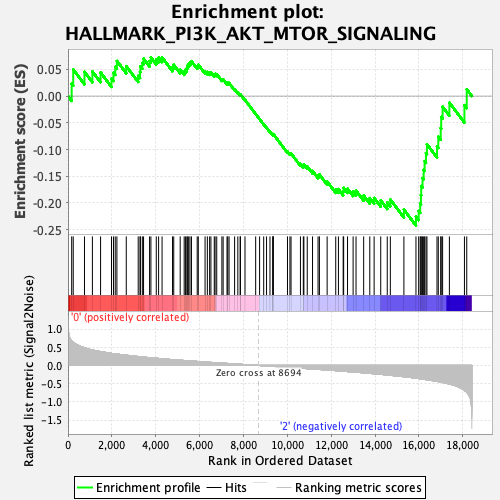
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group1_versus_Group3.Basophil_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_PI3K_AKT_MTOR_SIGNALING |
| Enrichment Score (ES) | -0.2423917 |
| Normalized Enrichment Score (NES) | -1.1851091 |
| Nominal p-value | 0.19444445 |
| FDR q-value | 0.68549305 |
| FWER p-Value | 0.92 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Sfn | 168 | 0.670 | 0.0231 | No |
| 2 | Prkaa2 | 238 | 0.633 | 0.0498 | No |
| 3 | Mknk1 | 751 | 0.489 | 0.0454 | No |
| 4 | Mapk10 | 1112 | 0.422 | 0.0461 | No |
| 5 | Cdk2 | 1485 | 0.382 | 0.0441 | No |
| 6 | Cltc | 1983 | 0.331 | 0.0329 | No |
| 7 | Ppp2r1b | 2073 | 0.325 | 0.0437 | No |
| 8 | Plcb1 | 2153 | 0.319 | 0.0548 | No |
| 9 | Pin1 | 2229 | 0.313 | 0.0657 | No |
| 10 | Sla | 2656 | 0.280 | 0.0560 | No |
| 11 | Akt1 | 3200 | 0.244 | 0.0381 | No |
| 12 | Rptor | 3283 | 0.239 | 0.0451 | No |
| 13 | Pikfyve | 3300 | 0.237 | 0.0556 | No |
| 14 | Arf1 | 3388 | 0.231 | 0.0620 | No |
| 15 | Fasl | 3444 | 0.228 | 0.0700 | No |
| 16 | Lck | 3719 | 0.214 | 0.0654 | No |
| 17 | Hsp90b1 | 3779 | 0.211 | 0.0723 | No |
| 18 | Grb2 | 4024 | 0.198 | 0.0685 | No |
| 19 | Akt1s1 | 4130 | 0.192 | 0.0720 | No |
| 20 | Slc2a1 | 4288 | 0.184 | 0.0723 | No |
| 21 | Gsk3b | 4765 | 0.159 | 0.0539 | No |
| 22 | Plcg1 | 4820 | 0.156 | 0.0585 | No |
| 23 | Tiam1 | 5114 | 0.144 | 0.0494 | No |
| 24 | Tnfrsf1a | 5296 | 0.135 | 0.0460 | No |
| 25 | Tsc2 | 5353 | 0.133 | 0.0494 | No |
| 26 | Ralb | 5419 | 0.131 | 0.0521 | No |
| 27 | Tbk1 | 5442 | 0.129 | 0.0571 | No |
| 28 | Actr2 | 5500 | 0.127 | 0.0601 | No |
| 29 | Map2k3 | 5560 | 0.124 | 0.0629 | No |
| 30 | Cdkn1b | 5628 | 0.122 | 0.0651 | No |
| 31 | Mapk9 | 5890 | 0.110 | 0.0561 | No |
| 32 | Cxcr4 | 5940 | 0.108 | 0.0586 | No |
| 33 | Mapk1 | 6248 | 0.094 | 0.0464 | No |
| 34 | Traf2 | 6352 | 0.089 | 0.0451 | No |
| 35 | Vav3 | 6456 | 0.085 | 0.0436 | No |
| 36 | Raf1 | 6515 | 0.083 | 0.0444 | No |
| 37 | Map3k7 | 6667 | 0.078 | 0.0399 | No |
| 38 | Myd88 | 6705 | 0.076 | 0.0415 | No |
| 39 | Ube2n | 6780 | 0.073 | 0.0410 | No |
| 40 | Ripk1 | 7015 | 0.065 | 0.0314 | No |
| 41 | Pten | 7072 | 0.063 | 0.0313 | No |
| 42 | Prkar2a | 7249 | 0.055 | 0.0244 | No |
| 43 | Mapk8 | 7283 | 0.054 | 0.0252 | No |
| 44 | Ptpn11 | 7342 | 0.051 | 0.0245 | No |
| 45 | Atf1 | 7595 | 0.041 | 0.0127 | No |
| 46 | Ywhab | 7742 | 0.035 | 0.0064 | No |
| 47 | Smad2 | 7848 | 0.031 | 0.0022 | No |
| 48 | Eif4e | 7860 | 0.031 | 0.0031 | No |
| 49 | Stat2 | 8068 | 0.022 | -0.0072 | No |
| 50 | Cab39l | 8555 | 0.005 | -0.0335 | No |
| 51 | Grk2 | 8734 | -0.001 | -0.0432 | No |
| 52 | Prkag1 | 8921 | -0.009 | -0.0529 | No |
| 53 | Arpc3 | 9051 | -0.015 | -0.0592 | No |
| 54 | Ube2d3 | 9207 | -0.021 | -0.0667 | No |
| 55 | Mknk2 | 9337 | -0.026 | -0.0725 | No |
| 56 | Dapp1 | 9340 | -0.026 | -0.0714 | No |
| 57 | Actr3 | 9361 | -0.026 | -0.0712 | No |
| 58 | Mapkap1 | 10005 | -0.051 | -0.1039 | No |
| 59 | Itpr2 | 10117 | -0.056 | -0.1073 | No |
| 60 | E2f1 | 10167 | -0.058 | -0.1072 | No |
| 61 | Calr | 10596 | -0.075 | -0.1269 | No |
| 62 | Irak4 | 10727 | -0.081 | -0.1301 | No |
| 63 | Acaca | 10753 | -0.081 | -0.1276 | No |
| 64 | Egfr | 10906 | -0.087 | -0.1317 | No |
| 65 | Pdk1 | 11147 | -0.097 | -0.1401 | No |
| 66 | Cab39 | 11399 | -0.107 | -0.1486 | No |
| 67 | Hras | 11461 | -0.110 | -0.1467 | No |
| 68 | Pak4 | 11813 | -0.124 | -0.1599 | No |
| 69 | Nck1 | 12205 | -0.141 | -0.1745 | No |
| 70 | Prkcb | 12328 | -0.147 | -0.1741 | No |
| 71 | Map2k6 | 12544 | -0.157 | -0.1782 | No |
| 72 | Arhgdia | 12566 | -0.158 | -0.1718 | No |
| 73 | Rps6ka1 | 12742 | -0.167 | -0.1733 | No |
| 74 | Gna14 | 12997 | -0.178 | -0.1786 | No |
| 75 | Ecsit | 13130 | -0.184 | -0.1769 | No |
| 76 | Pfn1 | 13478 | -0.203 | -0.1861 | No |
| 77 | Them4 | 13757 | -0.217 | -0.1908 | No |
| 78 | Pla2g12a | 13955 | -0.228 | -0.1906 | No |
| 79 | Dusp3 | 14256 | -0.243 | -0.1953 | No |
| 80 | Cdkn1a | 14553 | -0.261 | -0.1989 | No |
| 81 | Ddit3 | 14694 | -0.268 | -0.1936 | No |
| 82 | Nfkbib | 15312 | -0.309 | -0.2124 | No |
| 83 | Pitx2 | 15862 | -0.349 | -0.2256 | Yes |
| 84 | Cdk4 | 15985 | -0.359 | -0.2149 | Yes |
| 85 | Il2rg | 16055 | -0.365 | -0.2012 | Yes |
| 86 | Rps6ka3 | 16089 | -0.368 | -0.1852 | Yes |
| 87 | Sqstm1 | 16102 | -0.369 | -0.1681 | Yes |
| 88 | Trib3 | 16163 | -0.372 | -0.1535 | Yes |
| 89 | Rac1 | 16212 | -0.377 | -0.1380 | Yes |
| 90 | Cfl1 | 16256 | -0.381 | -0.1220 | Yes |
| 91 | Camk4 | 16319 | -0.388 | -0.1067 | Yes |
| 92 | Il4 | 16363 | -0.392 | -0.0901 | Yes |
| 93 | Ap2m1 | 16821 | -0.436 | -0.0941 | Yes |
| 94 | Nod1 | 16880 | -0.443 | -0.0759 | Yes |
| 95 | Rit1 | 16990 | -0.456 | -0.0599 | Yes |
| 96 | Pik3r3 | 17013 | -0.459 | -0.0390 | Yes |
| 97 | Cdk1 | 17077 | -0.467 | -0.0199 | Yes |
| 98 | Ppp1ca | 17386 | -0.507 | -0.0123 | Yes |
| 99 | Adcy2 | 18072 | -0.678 | -0.0170 | Yes |
| 100 | Csnk2b | 18177 | -0.734 | 0.0126 | Yes |

