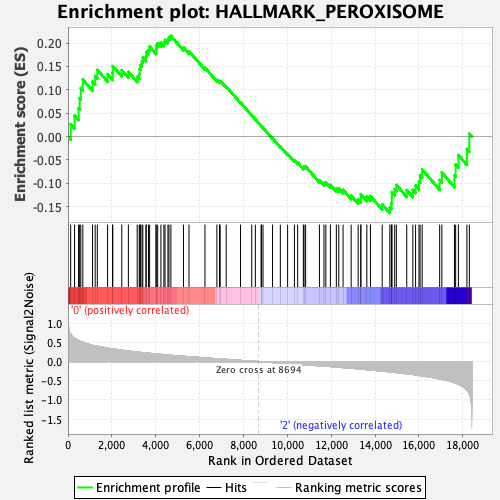
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group1_versus_Group3.Basophil_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 0 |
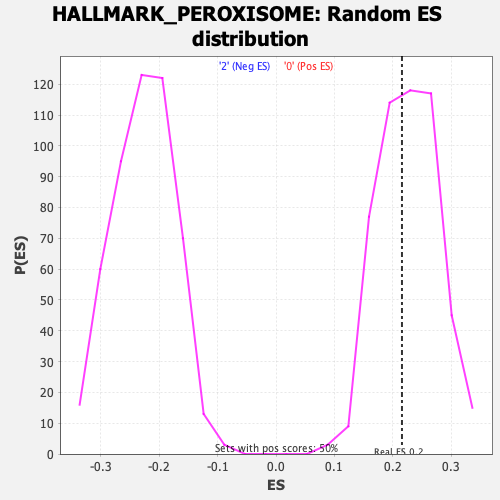
| GeneSet | HALLMARK_PEROXISOME |
| Enrichment Score (ES) | 0.21534044 |
| Normalized Enrichment Score (NES) | 0.952572 |
| Nominal p-value | 0.560241 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Pex6 | 122 | 0.713 | 0.0263 | Yes |
| 2 | Cln8 | 299 | 0.611 | 0.0449 | Yes |
| 3 | Ehhadh | 481 | 0.547 | 0.0603 | Yes |
| 4 | Abcb1a | 531 | 0.532 | 0.0823 | Yes |
| 5 | Idi1 | 583 | 0.522 | 0.1036 | Yes |
| 6 | Cacna1b | 675 | 0.504 | 0.1219 | Yes |
| 7 | Gnpat | 1119 | 0.422 | 0.1172 | Yes |
| 8 | Slc25a19 | 1238 | 0.408 | 0.1296 | Yes |
| 9 | Esr2 | 1337 | 0.397 | 0.1427 | Yes |
| 10 | Slc25a17 | 1804 | 0.348 | 0.1333 | Yes |
| 11 | Slc27a2 | 2031 | 0.327 | 0.1361 | Yes |
| 12 | Acox1 | 2041 | 0.327 | 0.1507 | Yes |
| 13 | Gstk1 | 2451 | 0.296 | 0.1420 | Yes |
| 14 | Hmgcl | 2752 | 0.275 | 0.1383 | Yes |
| 15 | Hsd17b4 | 3154 | 0.247 | 0.1279 | Yes |
| 16 | Fads1 | 3248 | 0.241 | 0.1340 | Yes |
| 17 | Abcb9 | 3266 | 0.241 | 0.1442 | Yes |
| 18 | Cnbp | 3299 | 0.237 | 0.1534 | Yes |
| 19 | Elovl5 | 3372 | 0.232 | 0.1602 | Yes |
| 20 | Mlycd | 3410 | 0.230 | 0.1688 | Yes |
| 21 | Aldh9a1 | 3555 | 0.222 | 0.1712 | Yes |
| 22 | Ywhah | 3564 | 0.222 | 0.1810 | Yes |
| 23 | Ercc3 | 3669 | 0.216 | 0.1853 | Yes |
| 24 | Fis1 | 3722 | 0.214 | 0.1924 | Yes |
| 25 | Dlg4 | 4013 | 0.198 | 0.1857 | Yes |
| 26 | Pabpc1 | 4019 | 0.198 | 0.1946 | Yes |
| 27 | Ide | 4087 | 0.194 | 0.1999 | Yes |
| 28 | Smarcc1 | 4236 | 0.186 | 0.2004 | Yes |
| 29 | Itgb1bp1 | 4371 | 0.179 | 0.2014 | Yes |
| 30 | Pex5 | 4423 | 0.176 | 0.2067 | Yes |
| 31 | Acsl4 | 4556 | 0.169 | 0.2074 | Yes |
| 32 | Abcb4 | 4603 | 0.167 | 0.2126 | Yes |
| 33 | Aldh1a1 | 4691 | 0.162 | 0.2153 | Yes |
| 34 | Vps4b | 5263 | 0.137 | 0.1905 | No |
| 35 | Msh2 | 5515 | 0.126 | 0.1826 | No |
| 36 | Slc23a2 | 6243 | 0.095 | 0.1473 | No |
| 37 | Isoc1 | 6787 | 0.073 | 0.1211 | No |
| 38 | Sema3c | 6905 | 0.069 | 0.1179 | No |
| 39 | Crat | 6938 | 0.067 | 0.1192 | No |
| 40 | Abcd1 | 7211 | 0.057 | 0.1070 | No |
| 41 | Sult2b1 | 7863 | 0.031 | 0.0729 | No |
| 42 | Rdh11 | 8376 | 0.011 | 0.0455 | No |
| 43 | Dhrs3 | 8540 | 0.005 | 0.0368 | No |
| 44 | Pex2 | 8802 | -0.004 | 0.0228 | No |
| 45 | Pex13 | 8823 | -0.005 | 0.0219 | No |
| 46 | Ercc1 | 8890 | -0.008 | 0.0187 | No |
| 47 | Sod2 | 9324 | -0.025 | -0.0038 | No |
| 48 | Siah1a | 9679 | -0.039 | -0.0213 | No |
| 49 | Abcd2 | 10011 | -0.051 | -0.0370 | No |
| 50 | Acsl5 | 10322 | -0.064 | -0.0510 | No |
| 51 | Slc35b2 | 10469 | -0.069 | -0.0558 | No |
| 52 | Atxn1 | 10728 | -0.081 | -0.0661 | No |
| 53 | Lonp2 | 10760 | -0.082 | -0.0641 | No |
| 54 | Ctbp1 | 10832 | -0.084 | -0.0640 | No |
| 55 | Hras | 11461 | -0.110 | -0.0933 | No |
| 56 | Acaa1a | 11668 | -0.118 | -0.0990 | No |
| 57 | Abcc5 | 11754 | -0.121 | -0.0981 | No |
| 58 | Hsd17b11 | 11962 | -0.130 | -0.1034 | No |
| 59 | Top2a | 12233 | -0.143 | -0.1115 | No |
| 60 | Hsd3b7 | 12343 | -0.147 | -0.1107 | No |
| 61 | Dhcr24 | 12539 | -0.157 | -0.1141 | No |
| 62 | Idh2 | 12908 | -0.175 | -0.1261 | No |
| 63 | Tspo | 13227 | -0.189 | -0.1347 | No |
| 64 | Pex14 | 13347 | -0.195 | -0.1322 | No |
| 65 | Cln6 | 13350 | -0.195 | -0.1233 | No |
| 66 | Cdk7 | 13623 | -0.210 | -0.1284 | No |
| 67 | Scp2 | 13788 | -0.218 | -0.1273 | No |
| 68 | Cadm1 | 14322 | -0.247 | -0.1449 | No |
| 69 | Idh1 | 14671 | -0.267 | -0.1516 | No |
| 70 | Retsat | 14739 | -0.271 | -0.1427 | No |
| 71 | Eci2 | 14761 | -0.272 | -0.1313 | No |
| 72 | Slc25a4 | 14764 | -0.273 | -0.1188 | No |
| 73 | Prdx1 | 14892 | -0.283 | -0.1127 | No |
| 74 | Mvp | 14968 | -0.288 | -0.1035 | No |
| 75 | Bcl10 | 15442 | -0.318 | -0.1146 | No |
| 76 | Abcd3 | 15720 | -0.337 | -0.1141 | No |
| 77 | Pex11b | 15845 | -0.347 | -0.1048 | No |
| 78 | Prdx5 | 15999 | -0.360 | -0.0965 | No |
| 79 | Acsl1 | 16050 | -0.364 | -0.0824 | No |
| 80 | Cat | 16145 | -0.372 | -0.0704 | No |
| 81 | Ephx2 | 16942 | -0.450 | -0.0930 | No |
| 82 | Ech1 | 17042 | -0.463 | -0.0770 | No |
| 83 | Nudt19 | 17616 | -0.548 | -0.0830 | No |
| 84 | Acot8 | 17671 | -0.559 | -0.0601 | No |
| 85 | Sod1 | 17796 | -0.588 | -0.0397 | No |
| 86 | Fdps | 18185 | -0.739 | -0.0267 | No |
| 87 | Pex11a | 18296 | -0.840 | 0.0061 | No |