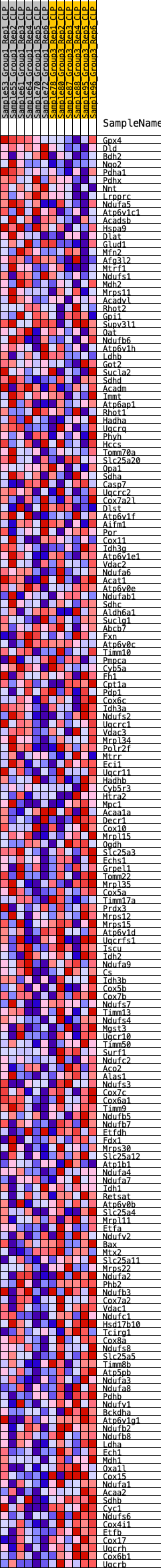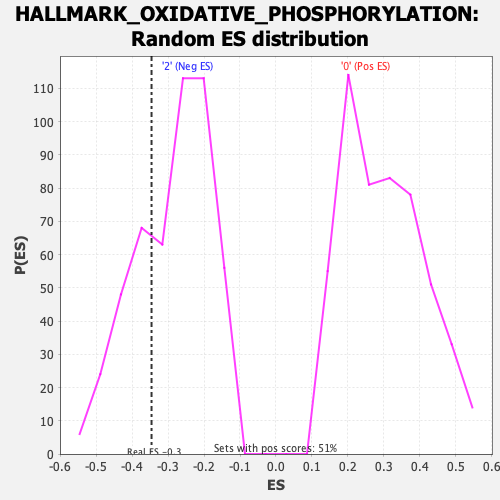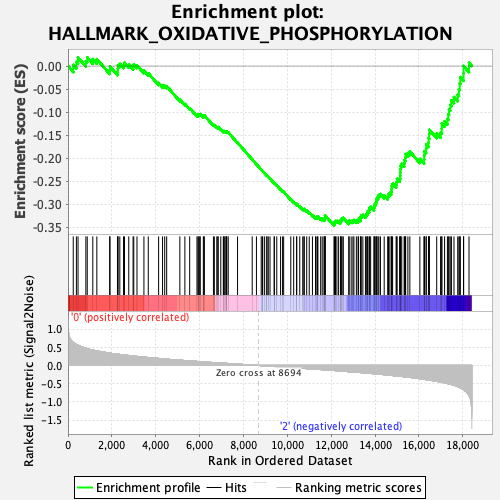
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group1_versus_Group3.Basophil_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_OXIDATIVE_PHOSPHORYLATION |
| Enrichment Score (ES) | -0.34593603 |
| Normalized Enrichment Score (NES) | -1.2029384 |
| Nominal p-value | 0.2790224 |
| FDR q-value | 0.78701776 |
| FWER p-Value | 0.908 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Gpx4 | 239 | 0.633 | 0.0024 | No |
| 2 | Dld | 382 | 0.578 | 0.0088 | No |
| 3 | Bdh2 | 457 | 0.557 | 0.0185 | No |
| 4 | Nqo2 | 814 | 0.478 | 0.0107 | No |
| 5 | Pdha1 | 877 | 0.465 | 0.0187 | No |
| 6 | Pdhx | 1131 | 0.420 | 0.0151 | No |
| 7 | Nnt | 1315 | 0.400 | 0.0149 | No |
| 8 | Lrpprc | 1900 | 0.339 | -0.0088 | No |
| 9 | Ndufa5 | 1915 | 0.337 | -0.0013 | No |
| 10 | Atp6v1c1 | 2255 | 0.311 | -0.0122 | No |
| 11 | Acadsb | 2273 | 0.310 | -0.0056 | No |
| 12 | Hspa9 | 2279 | 0.309 | 0.0018 | No |
| 13 | Dlat | 2362 | 0.302 | 0.0047 | No |
| 14 | Glud1 | 2531 | 0.289 | 0.0026 | No |
| 15 | Mfn2 | 2575 | 0.284 | 0.0072 | No |
| 16 | Afg3l2 | 2770 | 0.273 | 0.0033 | No |
| 17 | Mtrf1 | 2967 | 0.259 | -0.0011 | No |
| 18 | Ndufs1 | 2998 | 0.257 | 0.0035 | No |
| 19 | Mdh2 | 3145 | 0.248 | 0.0016 | No |
| 20 | Mrps11 | 3461 | 0.227 | -0.0101 | No |
| 21 | Acadvl | 3659 | 0.217 | -0.0156 | No |
| 22 | Rhot2 | 4131 | 0.192 | -0.0367 | No |
| 23 | Gpi1 | 4310 | 0.182 | -0.0420 | No |
| 24 | Supv3l1 | 4408 | 0.177 | -0.0430 | No |
| 25 | Oat | 4498 | 0.172 | -0.0436 | No |
| 26 | Ndufb6 | 5099 | 0.144 | -0.0730 | No |
| 27 | Atp6v1h | 5328 | 0.134 | -0.0822 | No |
| 28 | Ldhb | 5547 | 0.125 | -0.0911 | No |
| 29 | Got2 | 5879 | 0.110 | -0.1065 | No |
| 30 | Sucla2 | 5931 | 0.108 | -0.1067 | No |
| 31 | Sdhd | 5939 | 0.108 | -0.1044 | No |
| 32 | Acadm | 6001 | 0.105 | -0.1052 | No |
| 33 | Immt | 6029 | 0.105 | -0.1041 | No |
| 34 | Atp6ap1 | 6169 | 0.097 | -0.1093 | No |
| 35 | Rhot1 | 6197 | 0.096 | -0.1084 | No |
| 36 | Hadha | 6213 | 0.096 | -0.1069 | No |
| 37 | Uqcrq | 6630 | 0.079 | -0.1278 | No |
| 38 | Phyh | 6671 | 0.078 | -0.1281 | No |
| 39 | Hccs | 6783 | 0.073 | -0.1324 | No |
| 40 | Tomm70a | 6830 | 0.071 | -0.1331 | No |
| 41 | Slc25a20 | 6846 | 0.071 | -0.1322 | No |
| 42 | Opa1 | 6965 | 0.067 | -0.1370 | No |
| 43 | Sdha | 7080 | 0.063 | -0.1418 | No |
| 44 | Casp7 | 7093 | 0.062 | -0.1409 | No |
| 45 | Uqcrc2 | 7123 | 0.061 | -0.1410 | No |
| 46 | Cox7a2l | 7167 | 0.059 | -0.1419 | No |
| 47 | Dlst | 7222 | 0.056 | -0.1435 | No |
| 48 | Atp6v1f | 7224 | 0.056 | -0.1422 | No |
| 49 | Aifm1 | 7239 | 0.056 | -0.1416 | No |
| 50 | Por | 7303 | 0.053 | -0.1437 | No |
| 51 | Cox11 | 7730 | 0.036 | -0.1662 | No |
| 52 | Idh3g | 8397 | 0.010 | -0.2025 | No |
| 53 | Atp6v1e1 | 8590 | 0.004 | -0.2129 | No |
| 54 | Vdac2 | 8808 | -0.005 | -0.2247 | No |
| 55 | Ndufa6 | 8861 | -0.007 | -0.2274 | No |
| 56 | Acat1 | 8878 | -0.008 | -0.2281 | No |
| 57 | Atp6v0e | 8963 | -0.011 | -0.2324 | No |
| 58 | Ndufab1 | 9065 | -0.015 | -0.2376 | No |
| 59 | Sdhc | 9066 | -0.015 | -0.2372 | No |
| 60 | Aldh6a1 | 9130 | -0.018 | -0.2402 | No |
| 61 | Suclg1 | 9209 | -0.021 | -0.2440 | No |
| 62 | Abcb7 | 9398 | -0.028 | -0.2536 | No |
| 63 | Fxn | 9407 | -0.028 | -0.2534 | No |
| 64 | Atp6v0c | 9517 | -0.032 | -0.2586 | No |
| 65 | Timm10 | 9690 | -0.039 | -0.2671 | No |
| 66 | Pmpca | 9781 | -0.042 | -0.2710 | No |
| 67 | Cyb5a | 9831 | -0.044 | -0.2726 | No |
| 68 | Fh1 | 10156 | -0.057 | -0.2890 | No |
| 69 | Cpt1a | 10288 | -0.062 | -0.2946 | No |
| 70 | Pdp1 | 10421 | -0.067 | -0.3002 | No |
| 71 | Cox6c | 10426 | -0.067 | -0.2988 | No |
| 72 | Idh3a | 10564 | -0.074 | -0.3045 | No |
| 73 | Ndufs2 | 10701 | -0.079 | -0.3100 | No |
| 74 | Uqcrc1 | 10754 | -0.081 | -0.3108 | No |
| 75 | Vdac3 | 10770 | -0.082 | -0.3096 | No |
| 76 | Mrpl34 | 10882 | -0.086 | -0.3136 | No |
| 77 | Polr2f | 10997 | -0.090 | -0.3177 | No |
| 78 | Mtrr | 11143 | -0.097 | -0.3232 | No |
| 79 | Eci1 | 11282 | -0.103 | -0.3283 | No |
| 80 | Uqcr11 | 11311 | -0.104 | -0.3273 | No |
| 81 | Hadhb | 11379 | -0.107 | -0.3283 | No |
| 82 | Cyb5r3 | 11390 | -0.107 | -0.3262 | No |
| 83 | Htra2 | 11518 | -0.112 | -0.3305 | No |
| 84 | Mpc1 | 11588 | -0.115 | -0.3314 | No |
| 85 | Acaa1a | 11668 | -0.118 | -0.3329 | No |
| 86 | Decr1 | 11704 | -0.119 | -0.3319 | No |
| 87 | Cox10 | 11712 | -0.119 | -0.3293 | No |
| 88 | Mrpl15 | 11717 | -0.120 | -0.3266 | No |
| 89 | Ogdh | 11718 | -0.120 | -0.3237 | No |
| 90 | Slc25a3 | 12125 | -0.137 | -0.3426 | Yes |
| 91 | Echs1 | 12154 | -0.139 | -0.3407 | Yes |
| 92 | Grpel1 | 12163 | -0.139 | -0.3377 | Yes |
| 93 | Tomm22 | 12195 | -0.141 | -0.3360 | Yes |
| 94 | Mrpl35 | 12258 | -0.144 | -0.3358 | Yes |
| 95 | Cox5a | 12336 | -0.147 | -0.3365 | Yes |
| 96 | Timm17a | 12424 | -0.151 | -0.3375 | Yes |
| 97 | Prdx3 | 12430 | -0.152 | -0.3341 | Yes |
| 98 | Mrps12 | 12477 | -0.154 | -0.3328 | Yes |
| 99 | Mrps15 | 12491 | -0.155 | -0.3297 | Yes |
| 100 | Atp6v1d | 12550 | -0.157 | -0.3290 | Yes |
| 101 | Uqcrfs1 | 12799 | -0.170 | -0.3385 | Yes |
| 102 | Iscu | 12816 | -0.171 | -0.3351 | Yes |
| 103 | Idh2 | 12908 | -0.175 | -0.3358 | Yes |
| 104 | Ndufa9 | 12988 | -0.178 | -0.3358 | Yes |
| 105 | Cs | 13033 | -0.180 | -0.3338 | Yes |
| 106 | Idh3b | 13145 | -0.185 | -0.3353 | Yes |
| 107 | Cox5b | 13223 | -0.189 | -0.3349 | Yes |
| 108 | Cox7b | 13240 | -0.189 | -0.3312 | Yes |
| 109 | Ndufs7 | 13330 | -0.194 | -0.3313 | Yes |
| 110 | Timm13 | 13336 | -0.194 | -0.3268 | Yes |
| 111 | Ndufs4 | 13369 | -0.196 | -0.3237 | Yes |
| 112 | Mgst3 | 13441 | -0.201 | -0.3227 | Yes |
| 113 | Uqcr10 | 13559 | -0.207 | -0.3240 | Yes |
| 114 | Timm50 | 13598 | -0.209 | -0.3209 | Yes |
| 115 | Surf1 | 13649 | -0.211 | -0.3185 | Yes |
| 116 | Ndufc2 | 13670 | -0.212 | -0.3144 | Yes |
| 117 | Aco2 | 13734 | -0.216 | -0.3125 | Yes |
| 118 | Alas1 | 13740 | -0.216 | -0.3075 | Yes |
| 119 | Ndufs3 | 13796 | -0.219 | -0.3051 | Yes |
| 120 | Cox7c | 13947 | -0.228 | -0.3078 | Yes |
| 121 | Cox6a1 | 13961 | -0.229 | -0.3029 | Yes |
| 122 | Timm9 | 13992 | -0.230 | -0.2988 | Yes |
| 123 | Ndufb5 | 14037 | -0.233 | -0.2955 | Yes |
| 124 | Ndufb7 | 14056 | -0.233 | -0.2908 | Yes |
| 125 | Etfdh | 14082 | -0.235 | -0.2864 | Yes |
| 126 | Fdx1 | 14133 | -0.237 | -0.2833 | Yes |
| 127 | Mrps30 | 14172 | -0.238 | -0.2795 | Yes |
| 128 | Slc25a12 | 14243 | -0.242 | -0.2774 | Yes |
| 129 | Atp1b1 | 14414 | -0.252 | -0.2806 | Yes |
| 130 | Ndufa4 | 14573 | -0.262 | -0.2828 | Yes |
| 131 | Ndufa7 | 14609 | -0.264 | -0.2782 | Yes |
| 132 | Idh1 | 14671 | -0.267 | -0.2750 | Yes |
| 133 | Retsat | 14739 | -0.271 | -0.2721 | Yes |
| 134 | Atp6v0b | 14755 | -0.272 | -0.2662 | Yes |
| 135 | Slc25a4 | 14764 | -0.273 | -0.2599 | Yes |
| 136 | Mrpl11 | 14797 | -0.276 | -0.2549 | Yes |
| 137 | Etfa | 14957 | -0.287 | -0.2566 | Yes |
| 138 | Ndufv2 | 14986 | -0.289 | -0.2510 | Yes |
| 139 | Bax | 14999 | -0.289 | -0.2446 | Yes |
| 140 | Mtx2 | 15117 | -0.296 | -0.2437 | Yes |
| 141 | Slc25a11 | 15142 | -0.297 | -0.2378 | Yes |
| 142 | Mrps22 | 15148 | -0.297 | -0.2307 | Yes |
| 143 | Ndufa2 | 15155 | -0.298 | -0.2237 | Yes |
| 144 | Phb2 | 15165 | -0.298 | -0.2169 | Yes |
| 145 | Ndufb3 | 15214 | -0.301 | -0.2121 | Yes |
| 146 | Cox7a2 | 15323 | -0.310 | -0.2104 | Yes |
| 147 | Vdac1 | 15342 | -0.312 | -0.2038 | Yes |
| 148 | Ndufc1 | 15381 | -0.314 | -0.1981 | Yes |
| 149 | Hsd17b10 | 15392 | -0.315 | -0.1910 | Yes |
| 150 | Tcirg1 | 15494 | -0.322 | -0.1886 | Yes |
| 151 | Cox8a | 15584 | -0.328 | -0.1854 | Yes |
| 152 | Ndufs8 | 16040 | -0.364 | -0.2014 | Yes |
| 153 | Slc25a5 | 16231 | -0.378 | -0.2026 | Yes |
| 154 | Timm8b | 16240 | -0.379 | -0.1937 | Yes |
| 155 | Atp5pb | 16248 | -0.380 | -0.1847 | Yes |
| 156 | Ndufa3 | 16324 | -0.389 | -0.1793 | Yes |
| 157 | Ndufa8 | 16330 | -0.389 | -0.1700 | Yes |
| 158 | Pdhb | 16432 | -0.398 | -0.1658 | Yes |
| 159 | Ndufv1 | 16440 | -0.399 | -0.1564 | Yes |
| 160 | Bckdha | 16468 | -0.402 | -0.1480 | Yes |
| 161 | Atp6v1g1 | 16472 | -0.402 | -0.1382 | Yes |
| 162 | Ndufb2 | 16808 | -0.435 | -0.1459 | Yes |
| 163 | Ndufb8 | 16984 | -0.455 | -0.1444 | Yes |
| 164 | Ldha | 17037 | -0.462 | -0.1359 | Yes |
| 165 | Ech1 | 17042 | -0.463 | -0.1247 | Yes |
| 166 | Mdh1 | 17167 | -0.478 | -0.1198 | Yes |
| 167 | Oxa1l | 17296 | -0.495 | -0.1146 | Yes |
| 168 | Cox15 | 17338 | -0.502 | -0.1046 | Yes |
| 169 | Ndufa1 | 17369 | -0.506 | -0.0938 | Yes |
| 170 | Acaa2 | 17425 | -0.513 | -0.0842 | Yes |
| 171 | Sdhb | 17479 | -0.522 | -0.0743 | Yes |
| 172 | Cyc1 | 17595 | -0.545 | -0.0672 | Yes |
| 173 | Ndufs6 | 17766 | -0.579 | -0.0623 | Yes |
| 174 | Cox4i1 | 17819 | -0.595 | -0.0505 | Yes |
| 175 | Etfb | 17856 | -0.606 | -0.0376 | Yes |
| 176 | Cox17 | 17884 | -0.613 | -0.0240 | Yes |
| 177 | Uqcrh | 18031 | -0.658 | -0.0159 | Yes |
| 178 | Cox6b1 | 18035 | -0.660 | 0.0002 | Yes |
| 179 | Uqcrb | 18280 | -0.824 | 0.0070 | Yes |