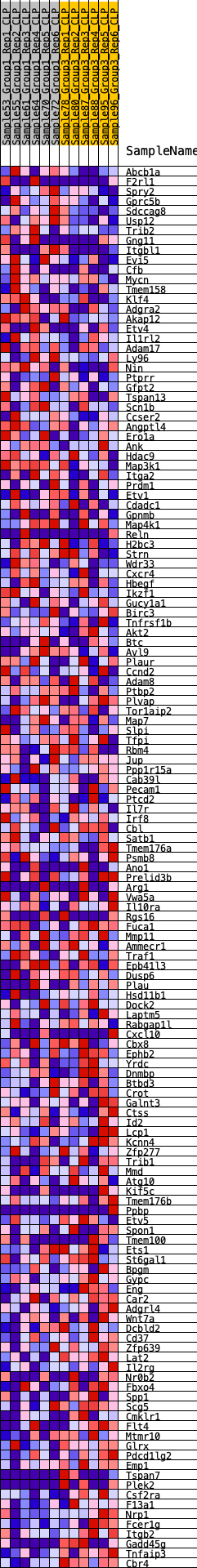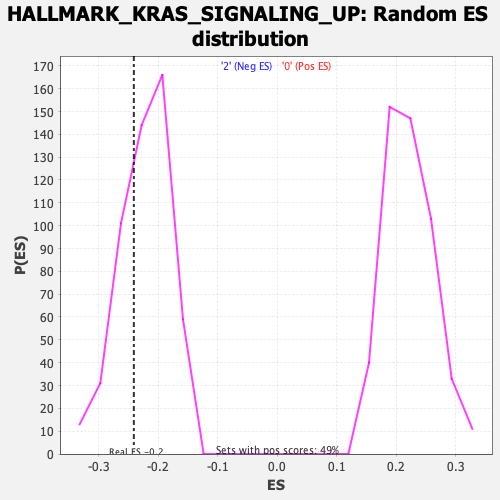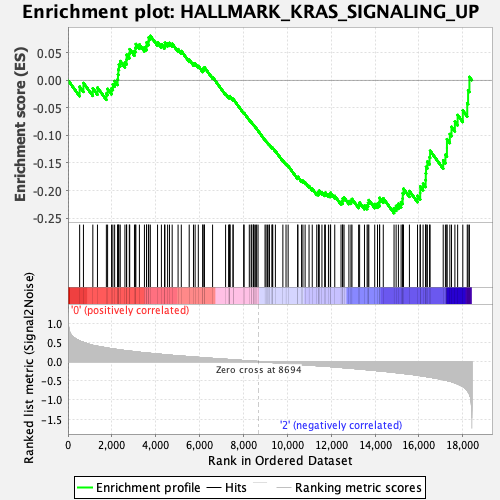
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group1_versus_Group3.Basophil_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_KRAS_SIGNALING_UP |
| Enrichment Score (ES) | -0.24103379 |
| Normalized Enrichment Score (NES) | -1.0833014 |
| Nominal p-value | 0.3385214 |
| FDR q-value | 0.6637862 |
| FWER p-Value | 0.978 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Abcb1a | 531 | 0.532 | -0.0118 | No |
| 2 | F2rl1 | 704 | 0.498 | -0.0050 | No |
| 3 | Spry2 | 1132 | 0.420 | -0.0148 | No |
| 4 | Gprc5b | 1343 | 0.396 | -0.0134 | No |
| 5 | Sdccag8 | 1749 | 0.355 | -0.0240 | No |
| 6 | Usp12 | 1805 | 0.348 | -0.0158 | No |
| 7 | Trib2 | 1985 | 0.331 | -0.0148 | No |
| 8 | Gng11 | 2045 | 0.326 | -0.0075 | No |
| 9 | Itgbl1 | 2126 | 0.321 | -0.0014 | No |
| 10 | Evi5 | 2260 | 0.311 | 0.0014 | No |
| 11 | Cfb | 2278 | 0.309 | 0.0105 | No |
| 12 | Mycn | 2296 | 0.307 | 0.0196 | No |
| 13 | Tmem158 | 2321 | 0.305 | 0.0282 | No |
| 14 | Klf4 | 2381 | 0.300 | 0.0347 | No |
| 15 | Adgra2 | 2595 | 0.284 | 0.0322 | No |
| 16 | Akap12 | 2667 | 0.280 | 0.0374 | No |
| 17 | Etv4 | 2674 | 0.280 | 0.0462 | No |
| 18 | Il1rl2 | 2795 | 0.271 | 0.0484 | No |
| 19 | Adam17 | 2813 | 0.270 | 0.0563 | No |
| 20 | Ly96 | 3030 | 0.255 | 0.0527 | No |
| 21 | Nin | 3070 | 0.252 | 0.0588 | No |
| 22 | Ptprr | 3094 | 0.251 | 0.0657 | No |
| 23 | Gfpt2 | 3255 | 0.241 | 0.0647 | No |
| 24 | Tspan13 | 3484 | 0.226 | 0.0596 | No |
| 25 | Scn1b | 3579 | 0.221 | 0.0616 | No |
| 26 | Ccser2 | 3584 | 0.221 | 0.0686 | No |
| 27 | Angptl4 | 3676 | 0.216 | 0.0706 | No |
| 28 | Ero1a | 3678 | 0.216 | 0.0776 | No |
| 29 | Ank | 3753 | 0.212 | 0.0804 | No |
| 30 | Hdac9 | 4082 | 0.195 | 0.0688 | No |
| 31 | Map3k1 | 4258 | 0.185 | 0.0652 | No |
| 32 | Itga2 | 4400 | 0.177 | 0.0632 | No |
| 33 | Prdm1 | 4419 | 0.176 | 0.0680 | No |
| 34 | Etv1 | 4541 | 0.170 | 0.0669 | No |
| 35 | Cdadc1 | 4626 | 0.166 | 0.0677 | No |
| 36 | Gpnmb | 4749 | 0.160 | 0.0662 | No |
| 37 | Map4k1 | 5018 | 0.148 | 0.0563 | No |
| 38 | Reln | 5165 | 0.141 | 0.0529 | No |
| 39 | H2bc3 | 5525 | 0.126 | 0.0374 | No |
| 40 | Strn | 5720 | 0.117 | 0.0305 | No |
| 41 | Wdr33 | 5795 | 0.114 | 0.0302 | No |
| 42 | Cxcr4 | 5940 | 0.108 | 0.0258 | No |
| 43 | Hbegf | 6142 | 0.099 | 0.0180 | No |
| 44 | Ikzf1 | 6149 | 0.098 | 0.0209 | No |
| 45 | Gucy1a1 | 6208 | 0.096 | 0.0208 | No |
| 46 | Birc3 | 6219 | 0.096 | 0.0234 | No |
| 47 | Tnfrsf1b | 6591 | 0.081 | 0.0057 | No |
| 48 | Akt2 | 7184 | 0.058 | -0.0248 | No |
| 49 | Btc | 7321 | 0.052 | -0.0306 | No |
| 50 | Avl9 | 7367 | 0.050 | -0.0314 | No |
| 51 | Plaur | 7381 | 0.050 | -0.0305 | No |
| 52 | Ccnd2 | 7382 | 0.050 | -0.0289 | No |
| 53 | Adam8 | 7525 | 0.044 | -0.0352 | No |
| 54 | Ptbp2 | 7526 | 0.044 | -0.0338 | No |
| 55 | Plvap | 8008 | 0.025 | -0.0593 | No |
| 56 | Tor1aip2 | 8028 | 0.024 | -0.0596 | No |
| 57 | Map7 | 8265 | 0.016 | -0.0720 | No |
| 58 | Slpi | 8348 | 0.012 | -0.0761 | No |
| 59 | Tfpi | 8375 | 0.011 | -0.0772 | No |
| 60 | Rbm4 | 8447 | 0.008 | -0.0808 | No |
| 61 | Jup | 8458 | 0.008 | -0.0811 | No |
| 62 | Ppp1r15a | 8487 | 0.007 | -0.0824 | No |
| 63 | Cab39l | 8555 | 0.005 | -0.0859 | No |
| 64 | Pecam1 | 8598 | 0.004 | -0.0881 | No |
| 65 | Ptcd2 | 8663 | 0.001 | -0.0916 | No |
| 66 | Il7r | 8982 | -0.012 | -0.1086 | No |
| 67 | Irf8 | 9029 | -0.014 | -0.1107 | No |
| 68 | Cbl | 9092 | -0.016 | -0.1135 | No |
| 69 | Satb1 | 9161 | -0.019 | -0.1166 | No |
| 70 | Tmem176a | 9183 | -0.020 | -0.1171 | No |
| 71 | Psmb8 | 9295 | -0.024 | -0.1224 | No |
| 72 | Ano1 | 9317 | -0.025 | -0.1228 | No |
| 73 | Prelid3b | 9334 | -0.025 | -0.1228 | No |
| 74 | Arg1 | 9454 | -0.030 | -0.1284 | No |
| 75 | Vwa5a | 9795 | -0.043 | -0.1456 | No |
| 76 | Il10ra | 9939 | -0.048 | -0.1519 | No |
| 77 | Rgs16 | 10035 | -0.052 | -0.1554 | No |
| 78 | Fuca1 | 10467 | -0.069 | -0.1767 | No |
| 79 | Mmp11 | 10481 | -0.070 | -0.1752 | No |
| 80 | Ammecr1 | 10647 | -0.077 | -0.1817 | No |
| 81 | Traf1 | 10709 | -0.080 | -0.1825 | No |
| 82 | Epb41l3 | 10809 | -0.084 | -0.1852 | No |
| 83 | Dusp6 | 10988 | -0.090 | -0.1920 | No |
| 84 | Plau | 11134 | -0.097 | -0.1968 | No |
| 85 | Hsd11b1 | 11339 | -0.105 | -0.2046 | No |
| 86 | Dock2 | 11414 | -0.108 | -0.2051 | No |
| 87 | Laptm5 | 11422 | -0.108 | -0.2020 | No |
| 88 | Rabgap1l | 11454 | -0.109 | -0.2001 | No |
| 89 | Cxcl10 | 11576 | -0.114 | -0.2031 | No |
| 90 | Cbx8 | 11700 | -0.119 | -0.2059 | No |
| 91 | Ephb2 | 11733 | -0.120 | -0.2038 | No |
| 92 | Yrdc | 11873 | -0.126 | -0.2073 | No |
| 93 | Dnmbp | 11958 | -0.130 | -0.2077 | No |
| 94 | Btbd3 | 11972 | -0.130 | -0.2042 | No |
| 95 | Crot | 12160 | -0.139 | -0.2099 | No |
| 96 | Galnt3 | 12446 | -0.152 | -0.2205 | No |
| 97 | Ctss | 12517 | -0.156 | -0.2193 | No |
| 98 | Id2 | 12523 | -0.156 | -0.2145 | No |
| 99 | Lcp1 | 12584 | -0.159 | -0.2126 | No |
| 100 | Kcnn4 | 12805 | -0.170 | -0.2191 | No |
| 101 | Zfp277 | 12899 | -0.174 | -0.2186 | No |
| 102 | Trib1 | 12945 | -0.176 | -0.2153 | No |
| 103 | Mmd | 13248 | -0.190 | -0.2257 | No |
| 104 | Atg10 | 13294 | -0.192 | -0.2219 | No |
| 105 | Kif5c | 13515 | -0.205 | -0.2273 | No |
| 106 | Tmem176b | 13636 | -0.210 | -0.2270 | No |
| 107 | Ppbp | 13679 | -0.213 | -0.2224 | No |
| 108 | Etv5 | 13714 | -0.215 | -0.2173 | No |
| 109 | Spon1 | 13982 | -0.230 | -0.2245 | No |
| 110 | Tmem100 | 14109 | -0.236 | -0.2237 | No |
| 111 | Ets1 | 14202 | -0.240 | -0.2209 | No |
| 112 | St6gal1 | 14207 | -0.240 | -0.2133 | No |
| 113 | Bpgm | 14372 | -0.250 | -0.2142 | No |
| 114 | Gypc | 14863 | -0.281 | -0.2319 | Yes |
| 115 | Eng | 14958 | -0.287 | -0.2277 | Yes |
| 116 | Car2 | 15062 | -0.293 | -0.2239 | Yes |
| 117 | Adgrl4 | 15185 | -0.300 | -0.2208 | Yes |
| 118 | Wnt7a | 15254 | -0.304 | -0.2146 | Yes |
| 119 | Dcbld2 | 15259 | -0.305 | -0.2049 | Yes |
| 120 | Cd37 | 15293 | -0.307 | -0.1968 | Yes |
| 121 | Zfp639 | 15564 | -0.326 | -0.2009 | Yes |
| 122 | Lat2 | 15933 | -0.354 | -0.2096 | Yes |
| 123 | Il2rg | 16055 | -0.365 | -0.2044 | Yes |
| 124 | Nr0b2 | 16057 | -0.365 | -0.1926 | Yes |
| 125 | Fbxo4 | 16187 | -0.374 | -0.1875 | Yes |
| 126 | Spp1 | 16302 | -0.387 | -0.1812 | Yes |
| 127 | Scg5 | 16310 | -0.387 | -0.1690 | Yes |
| 128 | Cmklr1 | 16322 | -0.388 | -0.1570 | Yes |
| 129 | Flt4 | 16382 | -0.394 | -0.1474 | Yes |
| 130 | Mtmr10 | 16482 | -0.403 | -0.1397 | Yes |
| 131 | Glrx | 16506 | -0.406 | -0.1278 | Yes |
| 132 | Pdcd1lg2 | 17104 | -0.471 | -0.1452 | Yes |
| 133 | Emp1 | 17205 | -0.482 | -0.1350 | Yes |
| 134 | Tspan7 | 17274 | -0.491 | -0.1228 | Yes |
| 135 | Plek2 | 17275 | -0.491 | -0.1069 | Yes |
| 136 | Csf2ra | 17407 | -0.511 | -0.0975 | Yes |
| 137 | F13a1 | 17483 | -0.523 | -0.0846 | Yes |
| 138 | Nrp1 | 17637 | -0.551 | -0.0750 | Yes |
| 139 | Fcer1g | 17762 | -0.578 | -0.0630 | Yes |
| 140 | Itgb2 | 17997 | -0.647 | -0.0548 | Yes |
| 141 | Gadd45g | 18200 | -0.753 | -0.0414 | Yes |
| 142 | Tnfaip3 | 18239 | -0.781 | -0.0182 | Yes |
| 143 | Cbr4 | 18299 | -0.842 | 0.0060 | Yes |