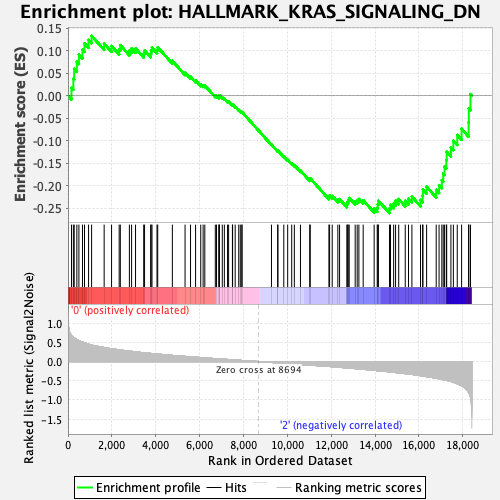
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group1_versus_Group3.Basophil_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_KRAS_SIGNALING_DN |
| Enrichment Score (ES) | -0.26108912 |
| Normalized Enrichment Score (NES) | -1.0354664 |
| Nominal p-value | 0.41082165 |
| FDR q-value | 0.6743507 |
| FWER p-Value | 0.992 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Rgs11 | 159 | 0.676 | 0.0176 | No |
| 2 | Cpeb3 | 244 | 0.630 | 0.0376 | No |
| 3 | Prodh | 282 | 0.618 | 0.0596 | No |
| 4 | Btg2 | 404 | 0.571 | 0.0752 | No |
| 5 | Vps50 | 497 | 0.542 | 0.0913 | No |
| 6 | Thnsl2 | 660 | 0.506 | 0.1021 | No |
| 7 | Nr4a2 | 752 | 0.489 | 0.1162 | No |
| 8 | Gpr19 | 938 | 0.450 | 0.1236 | No |
| 9 | Kcnn1 | 1076 | 0.427 | 0.1328 | No |
| 10 | Coq8a | 1648 | 0.365 | 0.1158 | No |
| 11 | Kcnd1 | 1986 | 0.330 | 0.1102 | No |
| 12 | Snn | 2333 | 0.304 | 0.1032 | No |
| 13 | Ngb | 2385 | 0.300 | 0.1121 | No |
| 14 | Sptbn2 | 2798 | 0.271 | 0.1001 | No |
| 15 | Plag1 | 2899 | 0.264 | 0.1049 | No |
| 16 | Magix | 3079 | 0.252 | 0.1050 | No |
| 17 | Nrip2 | 3451 | 0.228 | 0.0936 | No |
| 18 | Sidt1 | 3490 | 0.226 | 0.1003 | No |
| 19 | Clstn3 | 3768 | 0.211 | 0.0934 | No |
| 20 | Tg | 3788 | 0.210 | 0.1005 | No |
| 21 | Zc2hc1c | 3829 | 0.207 | 0.1064 | No |
| 22 | Slc29a3 | 4057 | 0.196 | 0.1016 | No |
| 23 | Nos1 | 4090 | 0.194 | 0.1074 | No |
| 24 | Htr1d | 4757 | 0.159 | 0.0773 | No |
| 25 | Kmt2d | 5336 | 0.133 | 0.0509 | No |
| 26 | Stag3 | 5586 | 0.123 | 0.0421 | No |
| 27 | Pdcd1 | 5816 | 0.113 | 0.0340 | No |
| 28 | Tgm1 | 6051 | 0.104 | 0.0252 | No |
| 29 | Tgfb2 | 6160 | 0.098 | 0.0232 | No |
| 30 | Dtnb | 6237 | 0.095 | 0.0227 | No |
| 31 | Capn9 | 6715 | 0.076 | -0.0004 | No |
| 32 | Dcc | 6771 | 0.074 | -0.0005 | No |
| 33 | Mfsd6 | 6881 | 0.069 | -0.0038 | No |
| 34 | Zbtb16 | 6888 | 0.069 | -0.0014 | No |
| 35 | Pde6b | 6899 | 0.069 | 0.0007 | No |
| 36 | Chst2 | 7033 | 0.064 | -0.0041 | No |
| 37 | Sgk1 | 7127 | 0.060 | -0.0068 | No |
| 38 | Nr6a1 | 7273 | 0.054 | -0.0126 | No |
| 39 | Prkn | 7322 | 0.052 | -0.0132 | No |
| 40 | Pdk2 | 7495 | 0.045 | -0.0208 | No |
| 41 | Itgb1bp2 | 7504 | 0.045 | -0.0195 | No |
| 42 | Brdt | 7621 | 0.040 | -0.0243 | No |
| 43 | Gp1ba | 7784 | 0.034 | -0.0318 | No |
| 44 | Atp4a | 7854 | 0.031 | -0.0344 | No |
| 45 | Ypel1 | 7926 | 0.028 | -0.0372 | No |
| 46 | Mx2 | 7933 | 0.028 | -0.0364 | No |
| 47 | Selenop | 9275 | -0.023 | -0.1088 | No |
| 48 | Thrb | 9558 | -0.034 | -0.1229 | No |
| 49 | Tfcp2l1 | 9570 | -0.034 | -0.1221 | No |
| 50 | Ptprj | 9837 | -0.044 | -0.1349 | No |
| 51 | Cd80 | 10014 | -0.051 | -0.1426 | No |
| 52 | Arpp21 | 10198 | -0.059 | -0.1503 | No |
| 53 | Cdkal1 | 10318 | -0.064 | -0.1543 | No |
| 54 | Lfng | 10597 | -0.075 | -0.1666 | No |
| 55 | Ryr2 | 11019 | -0.091 | -0.1860 | No |
| 56 | Tcf7l1 | 11042 | -0.092 | -0.1836 | No |
| 57 | Myo15a | 11894 | -0.127 | -0.2252 | No |
| 58 | Copz2 | 11925 | -0.128 | -0.2218 | No |
| 59 | Egf | 12044 | -0.133 | -0.2231 | No |
| 60 | Gamt | 12301 | -0.145 | -0.2314 | No |
| 61 | Epha5 | 12380 | -0.149 | -0.2299 | No |
| 62 | Bard1 | 12714 | -0.166 | -0.2416 | No |
| 63 | Celsr2 | 12733 | -0.167 | -0.2361 | No |
| 64 | Arhgdig | 12776 | -0.169 | -0.2318 | No |
| 65 | Cyp39a1 | 12818 | -0.171 | -0.2274 | No |
| 66 | Edar | 13091 | -0.183 | -0.2351 | No |
| 67 | Tenm2 | 13183 | -0.186 | -0.2328 | No |
| 68 | Mthfr | 13260 | -0.190 | -0.2296 | No |
| 69 | Msh5 | 13456 | -0.201 | -0.2324 | No |
| 70 | Ybx2 | 13956 | -0.228 | -0.2508 | No |
| 71 | Rsad2 | 14096 | -0.235 | -0.2492 | No |
| 72 | Gtf3c5 | 14122 | -0.237 | -0.2413 | No |
| 73 | Mast3 | 14149 | -0.238 | -0.2335 | No |
| 74 | Slc12a3 | 14655 | -0.265 | -0.2508 | Yes |
| 75 | Bmpr1b | 14702 | -0.269 | -0.2428 | Yes |
| 76 | Skil | 14845 | -0.280 | -0.2397 | Yes |
| 77 | Idua | 14933 | -0.285 | -0.2333 | Yes |
| 78 | Zfp112 | 15076 | -0.294 | -0.2297 | Yes |
| 79 | Ntf3 | 15373 | -0.314 | -0.2336 | Yes |
| 80 | Slc16a7 | 15522 | -0.324 | -0.2291 | Yes |
| 81 | Synpo | 15677 | -0.334 | -0.2245 | Yes |
| 82 | Ryr1 | 16070 | -0.366 | -0.2316 | Yes |
| 83 | Entpd7 | 16167 | -0.373 | -0.2224 | Yes |
| 84 | Camk1d | 16179 | -0.373 | -0.2084 | Yes |
| 85 | Tnni3 | 16348 | -0.392 | -0.2024 | Yes |
| 86 | Abcg4 | 16785 | -0.432 | -0.2094 | Yes |
| 87 | Efhd1 | 16915 | -0.446 | -0.1990 | Yes |
| 88 | Grid2 | 17041 | -0.463 | -0.1878 | Yes |
| 89 | Ccdc106 | 17105 | -0.471 | -0.1729 | Yes |
| 90 | Fggy | 17166 | -0.477 | -0.1576 | Yes |
| 91 | Asb7 | 17247 | -0.488 | -0.1430 | Yes |
| 92 | Mefv | 17263 | -0.489 | -0.1248 | Yes |
| 93 | Slc25a23 | 17455 | -0.518 | -0.1150 | Yes |
| 94 | Sphk2 | 17567 | -0.539 | -0.1001 | Yes |
| 95 | Klk8 | 17744 | -0.575 | -0.0874 | Yes |
| 96 | Tshb | 17943 | -0.631 | -0.0736 | Yes |
| 97 | Tex15 | 18263 | -0.805 | -0.0597 | Yes |
| 98 | Tent5c | 18271 | -0.815 | -0.0284 | Yes |
| 99 | Macroh2a2 | 18347 | -0.920 | 0.0033 | Yes |

