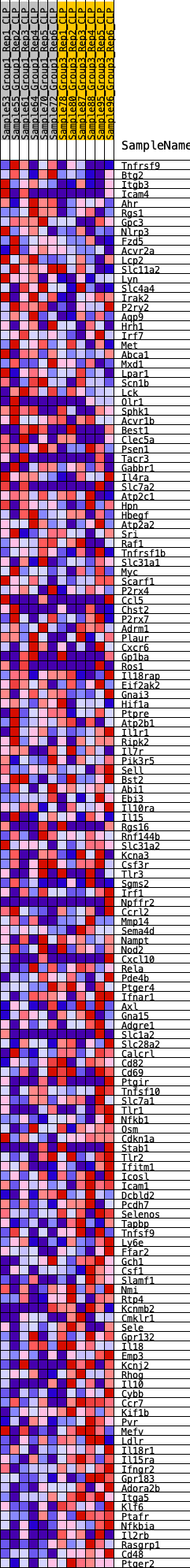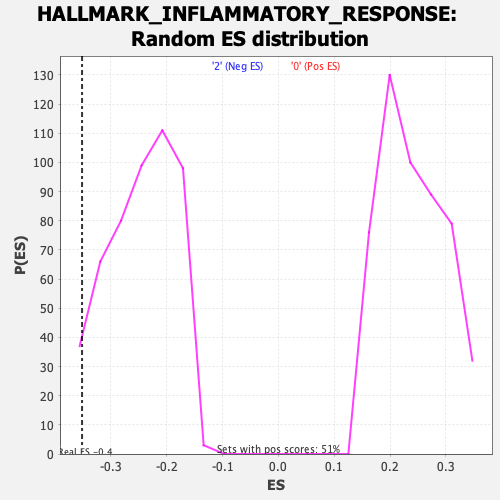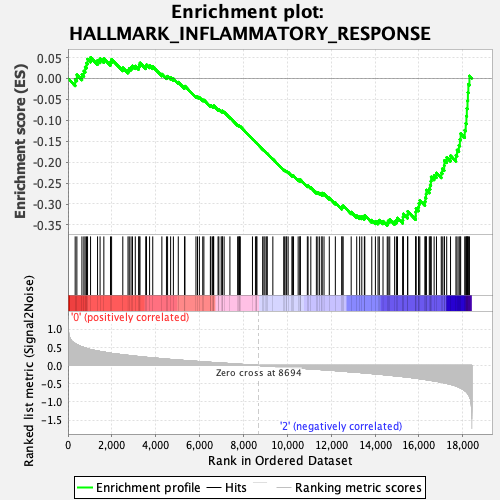
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group1_versus_Group3.Basophil_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_INFLAMMATORY_RESPONSE |
| Enrichment Score (ES) | -0.3515764 |
| Normalized Enrichment Score (NES) | -1.4298766 |
| Nominal p-value | 0.02631579 |
| FDR q-value | 0.58632207 |
| FWER p-Value | 0.543 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Tnfrsf9 | 327 | 0.597 | -0.0022 | No |
| 2 | Btg2 | 404 | 0.571 | 0.0087 | No |
| 3 | Itgb3 | 635 | 0.510 | 0.0096 | No |
| 4 | Icam4 | 724 | 0.493 | 0.0178 | No |
| 5 | Ahr | 782 | 0.483 | 0.0274 | No |
| 6 | Rgs1 | 842 | 0.471 | 0.0366 | No |
| 7 | Gpc3 | 891 | 0.462 | 0.0461 | No |
| 8 | Nlrp3 | 1028 | 0.435 | 0.0502 | No |
| 9 | Fzd5 | 1347 | 0.396 | 0.0432 | No |
| 10 | Acvr2a | 1462 | 0.384 | 0.0471 | No |
| 11 | Lcp2 | 1634 | 0.367 | 0.0474 | No |
| 12 | Slc11a2 | 1937 | 0.336 | 0.0397 | No |
| 13 | Lyn | 1987 | 0.330 | 0.0458 | No |
| 14 | Slc4a4 | 2498 | 0.292 | 0.0255 | No |
| 15 | Irak2 | 2736 | 0.276 | 0.0198 | No |
| 16 | P2ry2 | 2792 | 0.272 | 0.0240 | No |
| 17 | Aqp9 | 2874 | 0.266 | 0.0265 | No |
| 18 | Hrh1 | 2939 | 0.262 | 0.0299 | No |
| 19 | Irf7 | 3067 | 0.252 | 0.0296 | No |
| 20 | Met | 3221 | 0.243 | 0.0277 | No |
| 21 | Abca1 | 3246 | 0.242 | 0.0327 | No |
| 22 | Mxd1 | 3282 | 0.239 | 0.0371 | No |
| 23 | Lpar1 | 3544 | 0.223 | 0.0287 | No |
| 24 | Scn1b | 3579 | 0.221 | 0.0327 | No |
| 25 | Lck | 3719 | 0.214 | 0.0307 | No |
| 26 | Olr1 | 3863 | 0.206 | 0.0283 | No |
| 27 | Sphk1 | 4279 | 0.184 | 0.0104 | No |
| 28 | Acvr1b | 4488 | 0.173 | 0.0036 | No |
| 29 | Best1 | 4539 | 0.170 | 0.0054 | No |
| 30 | Clec5a | 4678 | 0.163 | 0.0021 | No |
| 31 | Psen1 | 4804 | 0.157 | -0.0006 | No |
| 32 | Tacr3 | 5027 | 0.147 | -0.0089 | No |
| 33 | Gabbr1 | 5310 | 0.134 | -0.0208 | No |
| 34 | Il4ra | 5332 | 0.133 | -0.0184 | No |
| 35 | Slc7a2 | 5833 | 0.112 | -0.0428 | No |
| 36 | Atp2c1 | 5903 | 0.109 | -0.0437 | No |
| 37 | Hpn | 5989 | 0.106 | -0.0456 | No |
| 38 | Hbegf | 6142 | 0.099 | -0.0513 | No |
| 39 | Atp2a2 | 6194 | 0.096 | -0.0515 | No |
| 40 | Sri | 6490 | 0.084 | -0.0655 | No |
| 41 | Raf1 | 6515 | 0.083 | -0.0646 | No |
| 42 | Tnfrsf1b | 6591 | 0.081 | -0.0666 | No |
| 43 | Slc31a1 | 6633 | 0.079 | -0.0668 | No |
| 44 | Myc | 6642 | 0.079 | -0.0651 | No |
| 45 | Scarf1 | 6841 | 0.071 | -0.0741 | No |
| 46 | P2rx4 | 6877 | 0.069 | -0.0742 | No |
| 47 | Ccl5 | 6983 | 0.066 | -0.0782 | No |
| 48 | Chst2 | 7033 | 0.064 | -0.0792 | No |
| 49 | P2rx7 | 7034 | 0.064 | -0.0775 | No |
| 50 | Adrm1 | 7117 | 0.061 | -0.0804 | No |
| 51 | Plaur | 7381 | 0.050 | -0.0935 | No |
| 52 | Cxcr6 | 7733 | 0.036 | -0.1117 | No |
| 53 | Gp1ba | 7784 | 0.034 | -0.1136 | No |
| 54 | Ros1 | 7797 | 0.033 | -0.1134 | No |
| 55 | Il18rap | 7817 | 0.033 | -0.1135 | No |
| 56 | Eif2ak2 | 7855 | 0.031 | -0.1148 | No |
| 57 | Gnai3 | 8409 | 0.010 | -0.1448 | No |
| 58 | Hif1a | 8545 | 0.005 | -0.1520 | No |
| 59 | Ptpre | 8603 | 0.003 | -0.1551 | No |
| 60 | Atp2b1 | 8607 | 0.003 | -0.1552 | No |
| 61 | Il1r1 | 8874 | -0.007 | -0.1695 | No |
| 62 | Ripk2 | 8932 | -0.010 | -0.1724 | No |
| 63 | Il7r | 8982 | -0.012 | -0.1748 | No |
| 64 | Pik3r5 | 9058 | -0.015 | -0.1785 | No |
| 65 | Sell | 9082 | -0.016 | -0.1793 | No |
| 66 | Bst2 | 9338 | -0.026 | -0.1926 | No |
| 67 | Abi1 | 9834 | -0.044 | -0.2185 | No |
| 68 | Ebi3 | 9878 | -0.046 | -0.2197 | No |
| 69 | Il10ra | 9939 | -0.048 | -0.2217 | No |
| 70 | Il15 | 9969 | -0.050 | -0.2220 | No |
| 71 | Rgs16 | 10035 | -0.052 | -0.2242 | No |
| 72 | Rnf144b | 10207 | -0.060 | -0.2320 | No |
| 73 | Slc31a2 | 10241 | -0.061 | -0.2322 | No |
| 74 | Kcna3 | 10281 | -0.062 | -0.2327 | No |
| 75 | Csf3r | 10492 | -0.070 | -0.2423 | No |
| 76 | Tlr3 | 10575 | -0.074 | -0.2449 | No |
| 77 | Sgms2 | 10580 | -0.074 | -0.2431 | No |
| 78 | Irf1 | 10584 | -0.075 | -0.2413 | No |
| 79 | Npffr2 | 10910 | -0.087 | -0.2568 | No |
| 80 | Ccrl2 | 10944 | -0.087 | -0.2563 | No |
| 81 | Mmp14 | 11069 | -0.093 | -0.2607 | No |
| 82 | Sema4d | 11322 | -0.104 | -0.2717 | No |
| 83 | Nampt | 11389 | -0.107 | -0.2725 | No |
| 84 | Nod2 | 11471 | -0.110 | -0.2740 | No |
| 85 | Cxcl10 | 11576 | -0.114 | -0.2767 | No |
| 86 | Rela | 11579 | -0.114 | -0.2738 | No |
| 87 | Pde4b | 11671 | -0.118 | -0.2757 | No |
| 88 | Ptger4 | 11911 | -0.128 | -0.2854 | No |
| 89 | Ifnar1 | 12187 | -0.140 | -0.2968 | No |
| 90 | Axl | 12480 | -0.154 | -0.3087 | No |
| 91 | Gna15 | 12495 | -0.155 | -0.3054 | No |
| 92 | Adgre1 | 12543 | -0.157 | -0.3038 | No |
| 93 | Slc1a2 | 12909 | -0.175 | -0.3192 | No |
| 94 | Slc28a2 | 13161 | -0.185 | -0.3281 | No |
| 95 | Calcrl | 13295 | -0.192 | -0.3303 | No |
| 96 | Cd82 | 13393 | -0.198 | -0.3304 | No |
| 97 | Cd69 | 13510 | -0.204 | -0.3313 | No |
| 98 | Ptgir | 13530 | -0.205 | -0.3270 | No |
| 99 | Tnfsf10 | 13848 | -0.222 | -0.3385 | No |
| 100 | Slc7a1 | 14013 | -0.231 | -0.3414 | No |
| 101 | Tlr1 | 14134 | -0.237 | -0.3417 | No |
| 102 | Nfkb1 | 14191 | -0.240 | -0.3384 | No |
| 103 | Osm | 14360 | -0.249 | -0.3411 | No |
| 104 | Cdkn1a | 14553 | -0.261 | -0.3447 | Yes |
| 105 | Stab1 | 14595 | -0.263 | -0.3400 | Yes |
| 106 | Tlr2 | 14667 | -0.266 | -0.3369 | Yes |
| 107 | Ifitm1 | 14890 | -0.283 | -0.3416 | Yes |
| 108 | Icosl | 14971 | -0.288 | -0.3384 | Yes |
| 109 | Icam1 | 15021 | -0.291 | -0.3334 | Yes |
| 110 | Dcbld2 | 15259 | -0.305 | -0.3383 | Yes |
| 111 | Pcdh7 | 15271 | -0.306 | -0.3309 | Yes |
| 112 | Selenos | 15287 | -0.307 | -0.3236 | Yes |
| 113 | Tapbp | 15482 | -0.321 | -0.3258 | Yes |
| 114 | Tnfsf9 | 15486 | -0.321 | -0.3175 | Yes |
| 115 | Ly6e | 15851 | -0.348 | -0.3282 | Yes |
| 116 | Ffar2 | 15852 | -0.348 | -0.3190 | Yes |
| 117 | Gch1 | 15865 | -0.349 | -0.3105 | Yes |
| 118 | Csf1 | 15982 | -0.359 | -0.3074 | Yes |
| 119 | Slamf1 | 15993 | -0.360 | -0.2984 | Yes |
| 120 | Nmi | 16038 | -0.364 | -0.2912 | Yes |
| 121 | Rtp4 | 16271 | -0.383 | -0.2938 | Yes |
| 122 | Kcnmb2 | 16290 | -0.386 | -0.2846 | Yes |
| 123 | Cmklr1 | 16322 | -0.388 | -0.2761 | Yes |
| 124 | Sele | 16336 | -0.390 | -0.2665 | Yes |
| 125 | Gpr132 | 16470 | -0.402 | -0.2632 | Yes |
| 126 | Il18 | 16509 | -0.406 | -0.2546 | Yes |
| 127 | Emp3 | 16541 | -0.410 | -0.2455 | Yes |
| 128 | Kcnj2 | 16554 | -0.412 | -0.2353 | Yes |
| 129 | Rhog | 16690 | -0.424 | -0.2315 | Yes |
| 130 | Il10 | 16797 | -0.434 | -0.2258 | Yes |
| 131 | Cybb | 17015 | -0.459 | -0.2256 | Yes |
| 132 | Ccr7 | 17060 | -0.465 | -0.2158 | Yes |
| 133 | Kif1b | 17149 | -0.476 | -0.2080 | Yes |
| 134 | Pvr | 17158 | -0.477 | -0.1959 | Yes |
| 135 | Mefv | 17263 | -0.489 | -0.1887 | Yes |
| 136 | Ldlr | 17438 | -0.515 | -0.1846 | Yes |
| 137 | Il18r1 | 17685 | -0.562 | -0.1833 | Yes |
| 138 | Il15ra | 17735 | -0.574 | -0.1708 | Yes |
| 139 | Ifngr2 | 17820 | -0.595 | -0.1597 | Yes |
| 140 | Gpr183 | 17864 | -0.608 | -0.1460 | Yes |
| 141 | Adora2b | 17900 | -0.619 | -0.1316 | Yes |
| 142 | Itga5 | 18086 | -0.687 | -0.1237 | Yes |
| 143 | Klf6 | 18131 | -0.708 | -0.1074 | Yes |
| 144 | Ptafr | 18162 | -0.724 | -0.0899 | Yes |
| 145 | Nfkbia | 18181 | -0.736 | -0.0715 | Yes |
| 146 | Il2rb | 18212 | -0.759 | -0.0531 | Yes |
| 147 | Rasgrp1 | 18230 | -0.773 | -0.0337 | Yes |
| 148 | Cd48 | 18251 | -0.796 | -0.0138 | Yes |
| 149 | Ptger2 | 18300 | -0.846 | 0.0059 | Yes |