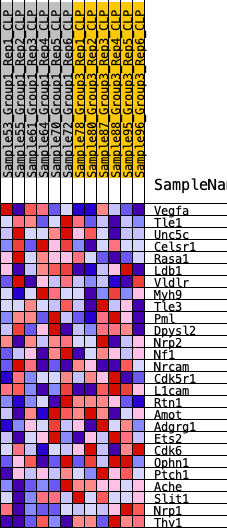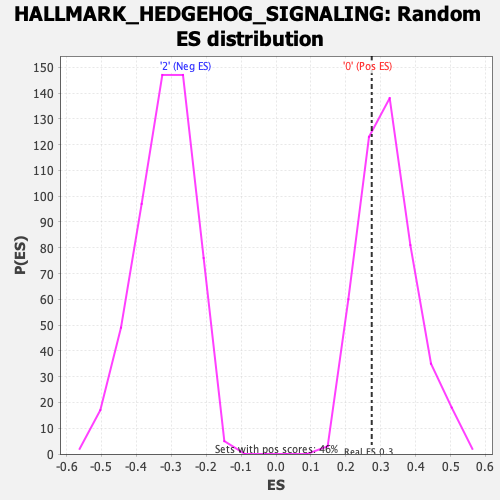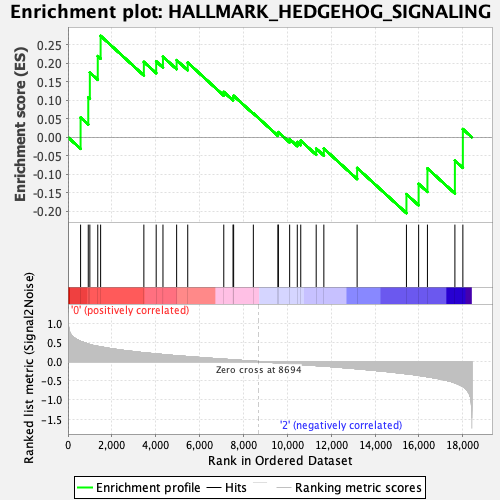
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group1_versus_Group3.Basophil_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_HEDGEHOG_SIGNALING |
| Enrichment Score (ES) | 0.27424493 |
| Normalized Enrichment Score (NES) | 0.8559928 |
| Nominal p-value | 0.6913043 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Vegfa | 574 | 0.524 | 0.0535 | Yes |
| 2 | Tle1 | 924 | 0.454 | 0.1080 | Yes |
| 3 | Unc5c | 996 | 0.440 | 0.1752 | Yes |
| 4 | Celsr1 | 1357 | 0.395 | 0.2195 | Yes |
| 5 | Rasa1 | 1487 | 0.382 | 0.2742 | Yes |
| 6 | Ldb1 | 3458 | 0.227 | 0.2039 | No |
| 7 | Vldlr | 4020 | 0.198 | 0.2054 | No |
| 8 | Myh9 | 4329 | 0.181 | 0.2179 | No |
| 9 | Tle3 | 4952 | 0.151 | 0.2086 | No |
| 10 | Pml | 5458 | 0.129 | 0.2020 | No |
| 11 | Dpysl2 | 7101 | 0.062 | 0.1226 | No |
| 12 | Nrp2 | 7523 | 0.044 | 0.1068 | No |
| 13 | Nf1 | 7551 | 0.043 | 0.1122 | No |
| 14 | Nrcam | 8449 | 0.008 | 0.0648 | No |
| 15 | Cdk5r1 | 9569 | -0.034 | 0.0094 | No |
| 16 | L1cam | 9590 | -0.035 | 0.0139 | No |
| 17 | Rtn1 | 10104 | -0.055 | -0.0051 | No |
| 18 | Amot | 10452 | -0.069 | -0.0128 | No |
| 19 | Adgrg1 | 10610 | -0.076 | -0.0091 | No |
| 20 | Ets2 | 11314 | -0.104 | -0.0306 | No |
| 21 | Cdk6 | 11663 | -0.117 | -0.0306 | No |
| 22 | Ophn1 | 13180 | -0.186 | -0.0830 | No |
| 23 | Ptch1 | 15428 | -0.317 | -0.1539 | No |
| 24 | Ache | 15984 | -0.359 | -0.1260 | No |
| 25 | Slit1 | 16385 | -0.394 | -0.0840 | No |
| 26 | Nrp1 | 17637 | -0.551 | -0.0629 | No |
| 27 | Thy1 | 18000 | -0.648 | 0.0222 | No |