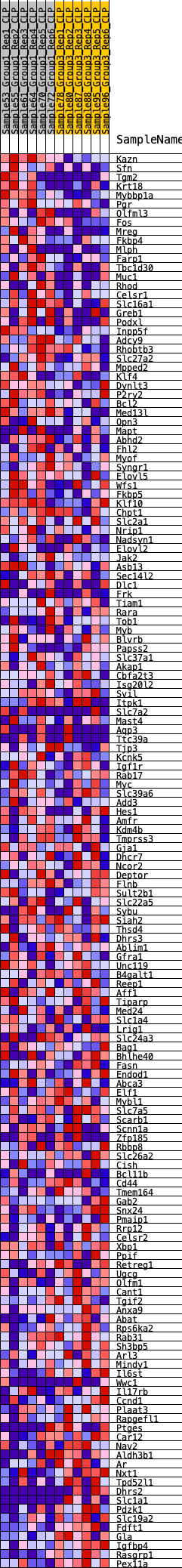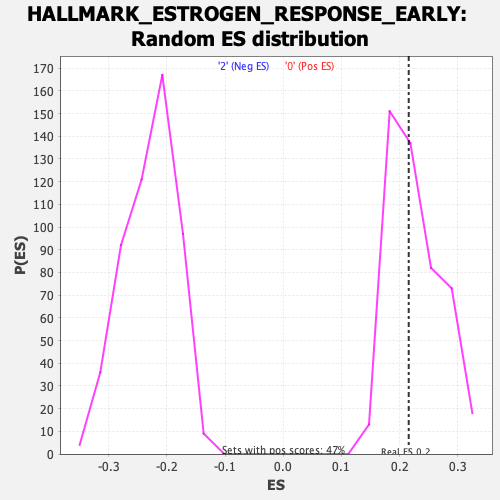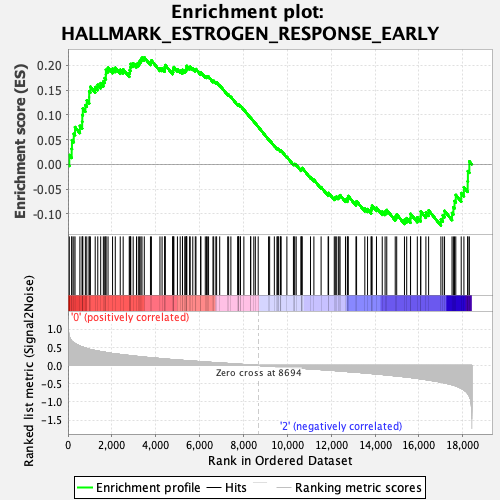
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group1_versus_Group3.Basophil_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_ESTROGEN_RESPONSE_EARLY |
| Enrichment Score (ES) | 0.21576965 |
| Normalized Enrichment Score (NES) | 0.95862603 |
| Nominal p-value | 0.48734176 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Kazn | 59 | 0.803 | 0.0189 | Yes |
| 2 | Sfn | 168 | 0.670 | 0.0315 | Yes |
| 3 | Tgm2 | 181 | 0.659 | 0.0490 | Yes |
| 4 | Krt18 | 264 | 0.623 | 0.0617 | Yes |
| 5 | Mybbp1a | 320 | 0.601 | 0.0753 | Yes |
| 6 | Pgr | 542 | 0.530 | 0.0778 | Yes |
| 7 | Olfml3 | 645 | 0.508 | 0.0862 | Yes |
| 8 | Fos | 652 | 0.507 | 0.0999 | Yes |
| 9 | Mreg | 676 | 0.503 | 0.1125 | Yes |
| 10 | Fkbp4 | 792 | 0.481 | 0.1195 | Yes |
| 11 | Mlph | 856 | 0.468 | 0.1290 | Yes |
| 12 | Farp1 | 965 | 0.446 | 0.1354 | Yes |
| 13 | Tbc1d30 | 966 | 0.446 | 0.1476 | Yes |
| 14 | Muc1 | 1021 | 0.436 | 0.1567 | Yes |
| 15 | Rhod | 1240 | 0.408 | 0.1560 | Yes |
| 16 | Celsr1 | 1357 | 0.395 | 0.1606 | Yes |
| 17 | Slc16a1 | 1490 | 0.381 | 0.1639 | Yes |
| 18 | Greb1 | 1612 | 0.369 | 0.1674 | Yes |
| 19 | Podxl | 1674 | 0.362 | 0.1741 | Yes |
| 20 | Inpp5f | 1726 | 0.357 | 0.1811 | Yes |
| 21 | Adcy9 | 1729 | 0.357 | 0.1908 | Yes |
| 22 | Rhobtb3 | 1825 | 0.346 | 0.1952 | Yes |
| 23 | Slc27a2 | 2031 | 0.327 | 0.1930 | Yes |
| 24 | Mpped2 | 2155 | 0.319 | 0.1950 | Yes |
| 25 | Klf4 | 2381 | 0.300 | 0.1910 | Yes |
| 26 | Dynlt3 | 2515 | 0.290 | 0.1917 | Yes |
| 27 | P2ry2 | 2792 | 0.272 | 0.1841 | Yes |
| 28 | Bcl2 | 2817 | 0.270 | 0.1902 | Yes |
| 29 | Med13l | 2848 | 0.268 | 0.1959 | Yes |
| 30 | Opn3 | 2861 | 0.267 | 0.2026 | Yes |
| 31 | Mapt | 2971 | 0.259 | 0.2038 | Yes |
| 32 | Abhd2 | 3122 | 0.249 | 0.2025 | Yes |
| 33 | Fhl2 | 3205 | 0.244 | 0.2047 | Yes |
| 34 | Myof | 3265 | 0.241 | 0.2081 | Yes |
| 35 | Syngr1 | 3321 | 0.236 | 0.2116 | Yes |
| 36 | Elovl5 | 3372 | 0.232 | 0.2153 | Yes |
| 37 | Wfs1 | 3478 | 0.227 | 0.2158 | Yes |
| 38 | Fkbp5 | 3759 | 0.212 | 0.2063 | No |
| 39 | Klf10 | 3802 | 0.209 | 0.2098 | No |
| 40 | Chpt1 | 4192 | 0.188 | 0.1936 | No |
| 41 | Slc2a1 | 4288 | 0.184 | 0.1935 | No |
| 42 | Nrip1 | 4407 | 0.177 | 0.1919 | No |
| 43 | Nadsyn1 | 4415 | 0.176 | 0.1964 | No |
| 44 | Elovl2 | 4432 | 0.175 | 0.2003 | No |
| 45 | Jak2 | 4768 | 0.159 | 0.1864 | No |
| 46 | Asb13 | 4790 | 0.158 | 0.1896 | No |
| 47 | Sec14l2 | 4801 | 0.157 | 0.1934 | No |
| 48 | Dlc1 | 4827 | 0.155 | 0.1963 | No |
| 49 | Frk | 4988 | 0.149 | 0.1916 | No |
| 50 | Tiam1 | 5114 | 0.144 | 0.1888 | No |
| 51 | Rara | 5218 | 0.139 | 0.1869 | No |
| 52 | Tob1 | 5220 | 0.139 | 0.1907 | No |
| 53 | Myb | 5324 | 0.134 | 0.1888 | No |
| 54 | Blvrb | 5367 | 0.132 | 0.1901 | No |
| 55 | Papss2 | 5393 | 0.132 | 0.1924 | No |
| 56 | Slc37a1 | 5408 | 0.131 | 0.1952 | No |
| 57 | Akap1 | 5412 | 0.131 | 0.1987 | No |
| 58 | Cbfa2t3 | 5546 | 0.125 | 0.1948 | No |
| 59 | Isg20l2 | 5568 | 0.124 | 0.1971 | No |
| 60 | Svil | 5688 | 0.119 | 0.1939 | No |
| 61 | Itpk1 | 5802 | 0.114 | 0.1908 | No |
| 62 | Slc7a2 | 5833 | 0.112 | 0.1923 | No |
| 63 | Mast4 | 6045 | 0.104 | 0.1836 | No |
| 64 | Aqp3 | 6056 | 0.104 | 0.1859 | No |
| 65 | Ttc39a | 6258 | 0.094 | 0.1775 | No |
| 66 | Tjp3 | 6316 | 0.091 | 0.1769 | No |
| 67 | Kcnk5 | 6348 | 0.090 | 0.1776 | No |
| 68 | Igf1r | 6408 | 0.087 | 0.1768 | No |
| 69 | Rab17 | 6609 | 0.080 | 0.1680 | No |
| 70 | Myc | 6642 | 0.079 | 0.1685 | No |
| 71 | Slc39a6 | 6741 | 0.075 | 0.1651 | No |
| 72 | Add3 | 6775 | 0.073 | 0.1654 | No |
| 73 | Hes1 | 6915 | 0.068 | 0.1596 | No |
| 74 | Amfr | 7281 | 0.054 | 0.1411 | No |
| 75 | Kdm4b | 7323 | 0.052 | 0.1403 | No |
| 76 | Tmprss3 | 7425 | 0.048 | 0.1361 | No |
| 77 | Gja1 | 7738 | 0.036 | 0.1200 | No |
| 78 | Dhcr7 | 7758 | 0.035 | 0.1199 | No |
| 79 | Ncor2 | 7760 | 0.035 | 0.1208 | No |
| 80 | Deptor | 7777 | 0.034 | 0.1209 | No |
| 81 | Flnb | 7850 | 0.031 | 0.1178 | No |
| 82 | Sult2b1 | 7863 | 0.031 | 0.1180 | No |
| 83 | Slc22a5 | 8017 | 0.025 | 0.1103 | No |
| 84 | Sybu | 8322 | 0.013 | 0.0940 | No |
| 85 | Siah2 | 8331 | 0.013 | 0.0939 | No |
| 86 | Thsd4 | 8470 | 0.008 | 0.0866 | No |
| 87 | Dhrs3 | 8540 | 0.005 | 0.0829 | No |
| 88 | Ablim1 | 8670 | 0.001 | 0.0759 | No |
| 89 | Gfra1 | 9143 | -0.018 | 0.0505 | No |
| 90 | Unc119 | 9192 | -0.020 | 0.0484 | No |
| 91 | B4galt1 | 9406 | -0.028 | 0.0375 | No |
| 92 | Reep1 | 9525 | -0.033 | 0.0320 | No |
| 93 | Aff1 | 9562 | -0.034 | 0.0309 | No |
| 94 | Tiparp | 9574 | -0.034 | 0.0313 | No |
| 95 | Med24 | 9606 | -0.035 | 0.0305 | No |
| 96 | Slc1a4 | 9688 | -0.039 | 0.0272 | No |
| 97 | Lrig1 | 9702 | -0.039 | 0.0276 | No |
| 98 | Slc24a3 | 9708 | -0.040 | 0.0284 | No |
| 99 | Bag1 | 9978 | -0.050 | 0.0150 | No |
| 100 | Bhlhe40 | 10276 | -0.062 | 0.0004 | No |
| 101 | Fasn | 10304 | -0.063 | 0.0007 | No |
| 102 | Endod1 | 10338 | -0.064 | 0.0006 | No |
| 103 | Abca3 | 10402 | -0.067 | -0.0010 | No |
| 104 | Elf1 | 10624 | -0.076 | -0.0110 | No |
| 105 | Mybl1 | 10628 | -0.076 | -0.0090 | No |
| 106 | Slc7a5 | 10657 | -0.077 | -0.0084 | No |
| 107 | Scarb1 | 10675 | -0.078 | -0.0072 | No |
| 108 | Scnn1a | 11055 | -0.093 | -0.0254 | No |
| 109 | Zfp185 | 11209 | -0.099 | -0.0311 | No |
| 110 | Rbbp8 | 11538 | -0.113 | -0.0459 | No |
| 111 | Slc26a2 | 11858 | -0.125 | -0.0600 | No |
| 112 | Cish | 11879 | -0.126 | -0.0576 | No |
| 113 | Bcl11b | 12138 | -0.138 | -0.0679 | No |
| 114 | Cd44 | 12184 | -0.140 | -0.0665 | No |
| 115 | Tmem164 | 12231 | -0.143 | -0.0651 | No |
| 116 | Gab2 | 12319 | -0.146 | -0.0658 | No |
| 117 | Snx24 | 12356 | -0.148 | -0.0637 | No |
| 118 | Pmaip1 | 12403 | -0.150 | -0.0621 | No |
| 119 | Rrp12 | 12649 | -0.163 | -0.0710 | No |
| 120 | Celsr2 | 12733 | -0.167 | -0.0709 | No |
| 121 | Xbp1 | 12756 | -0.168 | -0.0675 | No |
| 122 | Ppif | 12773 | -0.169 | -0.0637 | No |
| 123 | Retreg1 | 13127 | -0.184 | -0.0780 | No |
| 124 | Ugcg | 13155 | -0.185 | -0.0744 | No |
| 125 | Olfm1 | 13531 | -0.205 | -0.0892 | No |
| 126 | Cant1 | 13655 | -0.212 | -0.0901 | No |
| 127 | Tgif2 | 13819 | -0.220 | -0.0930 | No |
| 128 | Anxa9 | 13828 | -0.221 | -0.0874 | No |
| 129 | Abat | 13863 | -0.222 | -0.0831 | No |
| 130 | Rps6ka2 | 14062 | -0.234 | -0.0875 | No |
| 131 | Rab31 | 14325 | -0.248 | -0.0950 | No |
| 132 | Sh3bp5 | 14455 | -0.255 | -0.0950 | No |
| 133 | Arl3 | 14531 | -0.260 | -0.0920 | No |
| 134 | Mindy1 | 14917 | -0.284 | -0.1052 | No |
| 135 | Il6st | 14984 | -0.289 | -0.1009 | No |
| 136 | Wwc1 | 15338 | -0.311 | -0.1116 | No |
| 137 | Il17rb | 15440 | -0.318 | -0.1084 | No |
| 138 | Ccnd1 | 15612 | -0.330 | -0.1087 | No |
| 139 | Plaat3 | 15616 | -0.330 | -0.0997 | No |
| 140 | Rapgefl1 | 15924 | -0.354 | -0.1068 | No |
| 141 | Ptges | 16077 | -0.367 | -0.1050 | No |
| 142 | Car12 | 16079 | -0.367 | -0.0949 | No |
| 143 | Nav2 | 16315 | -0.388 | -0.0971 | No |
| 144 | Aldh3b1 | 16439 | -0.399 | -0.0928 | No |
| 145 | Ar | 16999 | -0.457 | -0.1108 | No |
| 146 | Nxt1 | 17085 | -0.468 | -0.1026 | No |
| 147 | Tpd52l1 | 17165 | -0.477 | -0.0937 | No |
| 148 | Dhrs2 | 17507 | -0.528 | -0.0979 | No |
| 149 | Slc1a1 | 17569 | -0.539 | -0.0863 | No |
| 150 | Pdzk1 | 17620 | -0.549 | -0.0739 | No |
| 151 | Slc19a2 | 17678 | -0.560 | -0.0616 | No |
| 152 | Fdft1 | 17925 | -0.626 | -0.0578 | No |
| 153 | Gla | 18048 | -0.668 | -0.0460 | No |
| 154 | Igfbp4 | 18219 | -0.764 | -0.0343 | No |
| 155 | Rasgrp1 | 18230 | -0.773 | -0.0135 | No |
| 156 | Pex11a | 18296 | -0.840 | 0.0061 | No |