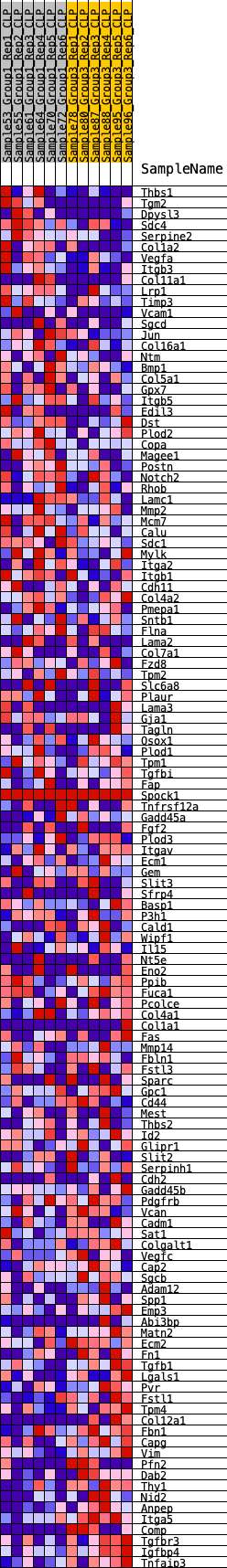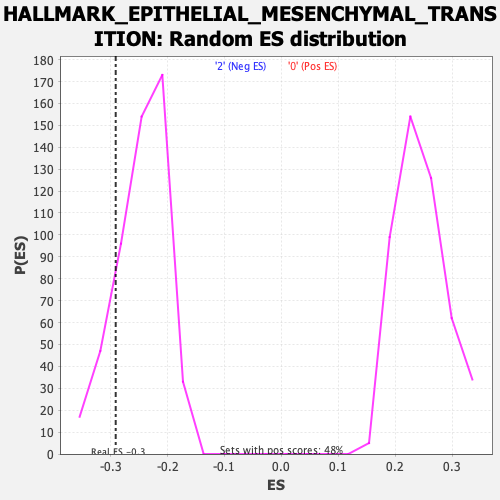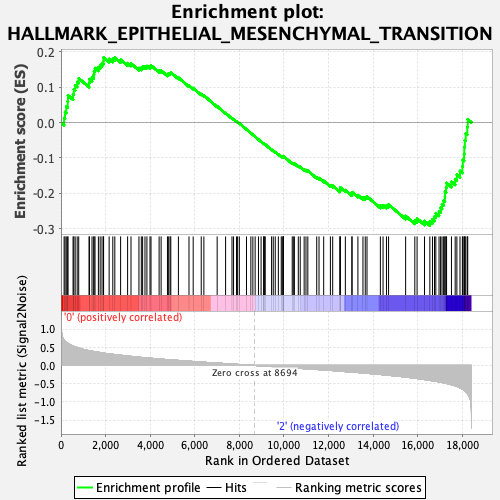
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group1_versus_Group3.Basophil_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION |
| Enrichment Score (ES) | -0.2911627 |
| Normalized Enrichment Score (NES) | -1.1804308 |
| Nominal p-value | 0.16346154 |
| FDR q-value | 0.63632536 |
| FWER p-Value | 0.926 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Thbs1 | 139 | 0.697 | 0.0128 | No |
| 2 | Tgm2 | 181 | 0.659 | 0.0298 | No |
| 3 | Dpysl3 | 237 | 0.633 | 0.0453 | No |
| 4 | Sdc4 | 300 | 0.609 | 0.0597 | No |
| 5 | Serpine2 | 312 | 0.604 | 0.0767 | No |
| 6 | Col1a2 | 541 | 0.530 | 0.0798 | No |
| 7 | Vegfa | 574 | 0.524 | 0.0933 | No |
| 8 | Itgb3 | 635 | 0.510 | 0.1050 | No |
| 9 | Col11a1 | 726 | 0.493 | 0.1144 | No |
| 10 | Lrp1 | 794 | 0.481 | 0.1248 | No |
| 11 | Timp3 | 1259 | 0.406 | 0.1113 | No |
| 12 | Vcam1 | 1274 | 0.405 | 0.1224 | No |
| 13 | Sgcd | 1391 | 0.392 | 0.1275 | No |
| 14 | Jun | 1468 | 0.383 | 0.1346 | No |
| 15 | Col16a1 | 1481 | 0.382 | 0.1451 | No |
| 16 | Ntm | 1526 | 0.377 | 0.1537 | No |
| 17 | Bmp1 | 1677 | 0.362 | 0.1561 | No |
| 18 | Col5a1 | 1756 | 0.355 | 0.1622 | No |
| 19 | Gpx7 | 1839 | 0.345 | 0.1678 | No |
| 20 | Itgb5 | 1907 | 0.338 | 0.1740 | No |
| 21 | Edil3 | 1909 | 0.338 | 0.1838 | No |
| 22 | Dst | 2159 | 0.318 | 0.1795 | No |
| 23 | Plod2 | 2323 | 0.305 | 0.1795 | No |
| 24 | Copa | 2413 | 0.298 | 0.1833 | No |
| 25 | Magee1 | 2673 | 0.280 | 0.1773 | No |
| 26 | Postn | 2987 | 0.258 | 0.1678 | No |
| 27 | Notch2 | 3139 | 0.249 | 0.1668 | No |
| 28 | Rhob | 3495 | 0.225 | 0.1539 | No |
| 29 | Lamc1 | 3602 | 0.220 | 0.1546 | No |
| 30 | Mmp2 | 3660 | 0.217 | 0.1578 | No |
| 31 | Mcm7 | 3766 | 0.212 | 0.1582 | No |
| 32 | Calu | 3853 | 0.206 | 0.1595 | No |
| 33 | Sdc1 | 3991 | 0.199 | 0.1579 | No |
| 34 | Mylk | 4036 | 0.197 | 0.1612 | No |
| 35 | Itga2 | 4400 | 0.177 | 0.1465 | No |
| 36 | Itgb1 | 4490 | 0.172 | 0.1467 | No |
| 37 | Cdh11 | 4774 | 0.158 | 0.1359 | No |
| 38 | Col4a2 | 4814 | 0.157 | 0.1383 | No |
| 39 | Pmepa1 | 4888 | 0.154 | 0.1388 | No |
| 40 | Sntb1 | 4924 | 0.153 | 0.1414 | No |
| 41 | Flna | 5265 | 0.137 | 0.1268 | No |
| 42 | Lama2 | 5740 | 0.116 | 0.1043 | No |
| 43 | Col7a1 | 5928 | 0.108 | 0.0972 | No |
| 44 | Fzd8 | 6289 | 0.092 | 0.0802 | No |
| 45 | Tpm2 | 6407 | 0.087 | 0.0763 | No |
| 46 | Slc6a8 | 7003 | 0.065 | 0.0457 | No |
| 47 | Plaur | 7381 | 0.050 | 0.0265 | No |
| 48 | Lama3 | 7668 | 0.038 | 0.0120 | No |
| 49 | Gja1 | 7738 | 0.036 | 0.0093 | No |
| 50 | Tagln | 7870 | 0.030 | 0.0030 | No |
| 51 | Qsox1 | 7920 | 0.029 | 0.0012 | No |
| 52 | Plod1 | 7998 | 0.026 | -0.0023 | No |
| 53 | Tpm1 | 8318 | 0.013 | -0.0194 | No |
| 54 | Tgfbi | 8514 | 0.006 | -0.0299 | No |
| 55 | Fap | 8604 | 0.003 | -0.0346 | No |
| 56 | Spock1 | 8706 | 0.000 | -0.0402 | No |
| 57 | Tnfrsf12a | 8856 | -0.007 | -0.0481 | No |
| 58 | Gadd45a | 8973 | -0.012 | -0.0541 | No |
| 59 | Fgf2 | 9088 | -0.016 | -0.0599 | No |
| 60 | Plod3 | 9098 | -0.017 | -0.0599 | No |
| 61 | Itgav | 9119 | -0.017 | -0.0605 | No |
| 62 | Ecm1 | 9159 | -0.019 | -0.0620 | No |
| 63 | Gem | 9444 | -0.029 | -0.0767 | No |
| 64 | Slit3 | 9518 | -0.032 | -0.0798 | No |
| 65 | Sfrp4 | 9608 | -0.035 | -0.0836 | No |
| 66 | Basp1 | 9747 | -0.041 | -0.0900 | No |
| 67 | P3h1 | 9877 | -0.046 | -0.0957 | No |
| 68 | Cald1 | 9921 | -0.048 | -0.0966 | No |
| 69 | Wipf1 | 9966 | -0.049 | -0.0976 | No |
| 70 | Il15 | 9969 | -0.050 | -0.0963 | No |
| 71 | Nt5e | 9973 | -0.050 | -0.0950 | No |
| 72 | Eno2 | 10369 | -0.065 | -0.1147 | No |
| 73 | Ppib | 10417 | -0.067 | -0.1153 | No |
| 74 | Fuca1 | 10467 | -0.069 | -0.1159 | No |
| 75 | Pcolce | 10642 | -0.077 | -0.1232 | No |
| 76 | Col4a1 | 10729 | -0.081 | -0.1256 | No |
| 77 | Col1a1 | 10904 | -0.087 | -0.1325 | No |
| 78 | Fas | 10986 | -0.090 | -0.1343 | No |
| 79 | Mmp14 | 11069 | -0.093 | -0.1361 | No |
| 80 | Fbln1 | 11468 | -0.110 | -0.1547 | No |
| 81 | Fstl3 | 11571 | -0.114 | -0.1569 | No |
| 82 | Sparc | 11780 | -0.123 | -0.1647 | No |
| 83 | Gpc1 | 12081 | -0.135 | -0.1772 | No |
| 84 | Cd44 | 12184 | -0.140 | -0.1787 | No |
| 85 | Mest | 12510 | -0.156 | -0.1919 | No |
| 86 | Thbs2 | 12519 | -0.156 | -0.1878 | No |
| 87 | Id2 | 12523 | -0.156 | -0.1834 | No |
| 88 | Glipr1 | 12752 | -0.168 | -0.1909 | No |
| 89 | Slit2 | 13044 | -0.181 | -0.2016 | No |
| 90 | Serpinh1 | 13053 | -0.181 | -0.1967 | No |
| 91 | Cdh2 | 13315 | -0.193 | -0.2053 | No |
| 92 | Gadd45b | 13541 | -0.206 | -0.2116 | No |
| 93 | Pdgfrb | 13644 | -0.211 | -0.2110 | No |
| 94 | Vcan | 13723 | -0.215 | -0.2090 | No |
| 95 | Cadm1 | 14322 | -0.247 | -0.2345 | No |
| 96 | Sat1 | 14442 | -0.254 | -0.2336 | No |
| 97 | Colgalt1 | 14596 | -0.263 | -0.2343 | No |
| 98 | Vegfc | 14690 | -0.268 | -0.2315 | No |
| 99 | Cap2 | 15455 | -0.318 | -0.2640 | No |
| 100 | Sgcb | 15867 | -0.350 | -0.2763 | No |
| 101 | Adam12 | 15969 | -0.358 | -0.2713 | No |
| 102 | Spp1 | 16302 | -0.387 | -0.2781 | No |
| 103 | Emp3 | 16541 | -0.410 | -0.2792 | Yes |
| 104 | Abi3bp | 16659 | -0.420 | -0.2733 | Yes |
| 105 | Matn2 | 16749 | -0.429 | -0.2656 | Yes |
| 106 | Ecm2 | 16809 | -0.435 | -0.2561 | Yes |
| 107 | Fn1 | 16953 | -0.452 | -0.2507 | Yes |
| 108 | Tgfb1 | 17023 | -0.460 | -0.2411 | Yes |
| 109 | Lgals1 | 17087 | -0.468 | -0.2308 | Yes |
| 110 | Pvr | 17158 | -0.477 | -0.2207 | Yes |
| 111 | Fstl1 | 17216 | -0.483 | -0.2097 | Yes |
| 112 | Tpm4 | 17220 | -0.483 | -0.1957 | Yes |
| 113 | Col12a1 | 17270 | -0.491 | -0.1841 | Yes |
| 114 | Fbn1 | 17291 | -0.494 | -0.1707 | Yes |
| 115 | Capg | 17510 | -0.528 | -0.1672 | Yes |
| 116 | Vim | 17677 | -0.560 | -0.1600 | Yes |
| 117 | Pfn2 | 17753 | -0.576 | -0.1472 | Yes |
| 118 | Dab2 | 17893 | -0.617 | -0.1368 | Yes |
| 119 | Thy1 | 18000 | -0.648 | -0.1237 | Yes |
| 120 | Nid2 | 18020 | -0.655 | -0.1056 | Yes |
| 121 | Anpep | 18078 | -0.680 | -0.0888 | Yes |
| 122 | Itga5 | 18086 | -0.687 | -0.0691 | Yes |
| 123 | Comp | 18110 | -0.698 | -0.0500 | Yes |
| 124 | Tgfbr3 | 18150 | -0.717 | -0.0312 | Yes |
| 125 | Igfbp4 | 18219 | -0.764 | -0.0125 | Yes |
| 126 | Tnfaip3 | 18239 | -0.781 | 0.0092 | Yes |