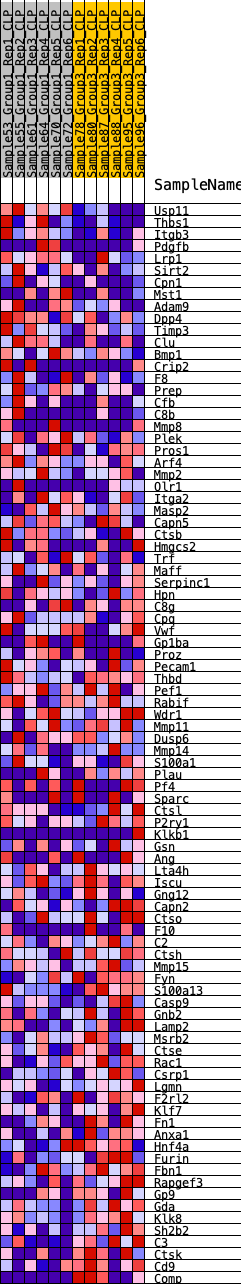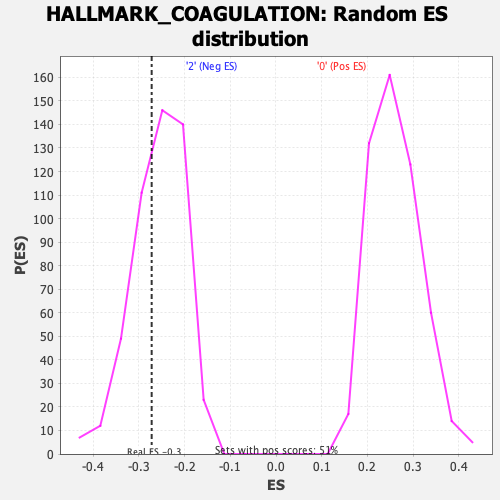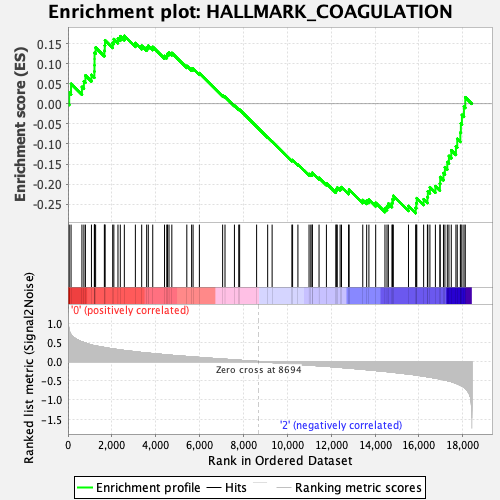
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group1_versus_Group3.Basophil_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_COAGULATION |
| Enrichment Score (ES) | -0.27220285 |
| Normalized Enrichment Score (NES) | -1.0604616 |
| Nominal p-value | 0.35655737 |
| FDR q-value | 0.6864044 |
| FWER p-Value | 0.984 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Usp11 | 55 | 0.810 | 0.0278 | No |
| 2 | Thbs1 | 139 | 0.697 | 0.0498 | No |
| 3 | Itgb3 | 635 | 0.510 | 0.0422 | No |
| 4 | Pdgfb | 725 | 0.493 | 0.0561 | No |
| 5 | Lrp1 | 794 | 0.481 | 0.0707 | No |
| 6 | Sirt2 | 1069 | 0.428 | 0.0720 | No |
| 7 | Cpn1 | 1201 | 0.413 | 0.0806 | No |
| 8 | Mst1 | 1207 | 0.412 | 0.0960 | No |
| 9 | Adam9 | 1210 | 0.412 | 0.1116 | No |
| 10 | Dpp4 | 1211 | 0.412 | 0.1272 | No |
| 11 | Timp3 | 1259 | 0.406 | 0.1401 | No |
| 12 | Clu | 1657 | 0.363 | 0.1323 | No |
| 13 | Bmp1 | 1677 | 0.362 | 0.1450 | No |
| 14 | Crip2 | 1688 | 0.361 | 0.1582 | No |
| 15 | F8 | 2037 | 0.327 | 0.1516 | No |
| 16 | Prep | 2090 | 0.323 | 0.1611 | No |
| 17 | Cfb | 2278 | 0.309 | 0.1626 | No |
| 18 | C8b | 2384 | 0.300 | 0.1683 | No |
| 19 | Mmp8 | 2568 | 0.285 | 0.1692 | No |
| 20 | Plek | 3071 | 0.252 | 0.1514 | No |
| 21 | Pros1 | 3359 | 0.233 | 0.1446 | No |
| 22 | Arf4 | 3586 | 0.221 | 0.1406 | No |
| 23 | Mmp2 | 3660 | 0.217 | 0.1449 | No |
| 24 | Olr1 | 3863 | 0.206 | 0.1417 | No |
| 25 | Itga2 | 4400 | 0.177 | 0.1192 | No |
| 26 | Masp2 | 4504 | 0.171 | 0.1201 | No |
| 27 | Capn5 | 4538 | 0.170 | 0.1247 | No |
| 28 | Ctsb | 4605 | 0.167 | 0.1275 | No |
| 29 | Hmgcs2 | 4733 | 0.160 | 0.1267 | No |
| 30 | Trf | 5416 | 0.131 | 0.0944 | No |
| 31 | Maff | 5635 | 0.121 | 0.0871 | No |
| 32 | Serpinc1 | 5702 | 0.118 | 0.0880 | No |
| 33 | Hpn | 5989 | 0.106 | 0.0764 | No |
| 34 | C8g | 7049 | 0.064 | 0.0210 | No |
| 35 | Cpq | 7157 | 0.059 | 0.0175 | No |
| 36 | Vwf | 7587 | 0.042 | -0.0044 | No |
| 37 | Gp1ba | 7784 | 0.034 | -0.0138 | No |
| 38 | Proz | 7824 | 0.032 | -0.0147 | No |
| 39 | Pecam1 | 8598 | 0.004 | -0.0568 | No |
| 40 | Thbd | 9102 | -0.017 | -0.0836 | No |
| 41 | Pef1 | 9305 | -0.024 | -0.0937 | No |
| 42 | Rabif | 10206 | -0.060 | -0.1405 | No |
| 43 | Wdr1 | 10238 | -0.061 | -0.1399 | No |
| 44 | Mmp11 | 10481 | -0.070 | -0.1505 | No |
| 45 | Dusp6 | 10988 | -0.090 | -0.1747 | No |
| 46 | Mmp14 | 11069 | -0.093 | -0.1755 | No |
| 47 | S100a1 | 11133 | -0.096 | -0.1753 | No |
| 48 | Plau | 11134 | -0.097 | -0.1716 | No |
| 49 | Pf4 | 11444 | -0.109 | -0.1843 | No |
| 50 | Sparc | 11780 | -0.123 | -0.1980 | No |
| 51 | Ctsl | 12209 | -0.141 | -0.2159 | No |
| 52 | P2ry1 | 12234 | -0.143 | -0.2118 | No |
| 53 | Klkb1 | 12275 | -0.144 | -0.2085 | No |
| 54 | Gsn | 12405 | -0.150 | -0.2098 | No |
| 55 | Ang | 12468 | -0.153 | -0.2074 | No |
| 56 | Lta4h | 12789 | -0.170 | -0.2184 | No |
| 57 | Iscu | 12816 | -0.171 | -0.2133 | No |
| 58 | Gng12 | 13437 | -0.200 | -0.2396 | No |
| 59 | Capn2 | 13615 | -0.210 | -0.2412 | No |
| 60 | Ctso | 13717 | -0.215 | -0.2386 | No |
| 61 | F10 | 14026 | -0.232 | -0.2466 | No |
| 62 | C2 | 14449 | -0.254 | -0.2599 | Yes |
| 63 | Ctsh | 14542 | -0.260 | -0.2551 | Yes |
| 64 | Mmp15 | 14604 | -0.263 | -0.2484 | Yes |
| 65 | Fyn | 14768 | -0.273 | -0.2469 | Yes |
| 66 | S100a13 | 14793 | -0.276 | -0.2377 | Yes |
| 67 | Casp9 | 14830 | -0.279 | -0.2291 | Yes |
| 68 | Gnb2 | 15521 | -0.324 | -0.2544 | Yes |
| 69 | Lamp2 | 15848 | -0.348 | -0.2590 | Yes |
| 70 | Msrb2 | 15878 | -0.350 | -0.2472 | Yes |
| 71 | Ctse | 15899 | -0.352 | -0.2349 | Yes |
| 72 | Rac1 | 16212 | -0.377 | -0.2376 | Yes |
| 73 | Csrp1 | 16384 | -0.394 | -0.2320 | Yes |
| 74 | Lgmn | 16404 | -0.395 | -0.2180 | Yes |
| 75 | F2rl2 | 16498 | -0.404 | -0.2077 | Yes |
| 76 | Klf7 | 16748 | -0.429 | -0.2050 | Yes |
| 77 | Fn1 | 16953 | -0.452 | -0.1989 | Yes |
| 78 | Anxa1 | 16966 | -0.453 | -0.1823 | Yes |
| 79 | Hnf4a | 17116 | -0.472 | -0.1725 | Yes |
| 80 | Furin | 17182 | -0.479 | -0.1578 | Yes |
| 81 | Fbn1 | 17291 | -0.494 | -0.1449 | Yes |
| 82 | Rapgef3 | 17365 | -0.505 | -0.1297 | Yes |
| 83 | Gp9 | 17472 | -0.520 | -0.1157 | Yes |
| 84 | Gda | 17684 | -0.562 | -0.1059 | Yes |
| 85 | Klk8 | 17744 | -0.575 | -0.0872 | Yes |
| 86 | Sh2b2 | 17885 | -0.614 | -0.0715 | Yes |
| 87 | C3 | 17910 | -0.622 | -0.0492 | Yes |
| 88 | Ctsk | 17958 | -0.636 | -0.0275 | Yes |
| 89 | Cd9 | 18050 | -0.669 | -0.0071 | Yes |
| 90 | Comp | 18110 | -0.698 | 0.0163 | Yes |