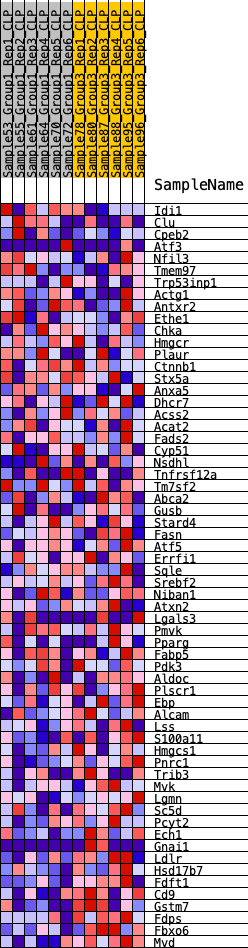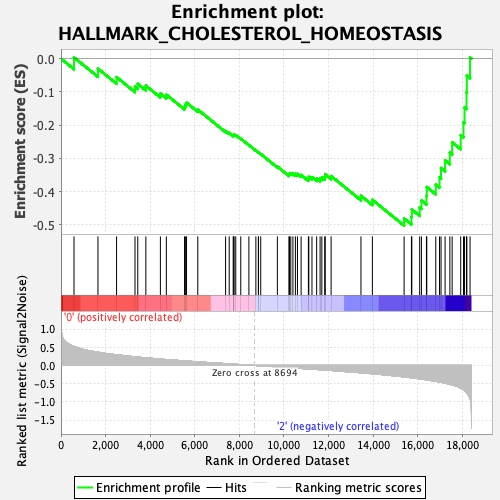
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group1_versus_Group3.Basophil_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_CHOLESTEROL_HOMEOSTASIS |
| Enrichment Score (ES) | -0.5022871 |
| Normalized Enrichment Score (NES) | -1.6807883 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.25164235 |
| FWER p-Value | 0.123 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Idi1 | 583 | 0.522 | 0.0037 | No |
| 2 | Clu | 1657 | 0.363 | -0.0301 | No |
| 3 | Cpeb2 | 2492 | 0.292 | -0.0557 | No |
| 4 | Atf3 | 3318 | 0.236 | -0.0846 | No |
| 5 | Nfil3 | 3443 | 0.228 | -0.0758 | No |
| 6 | Tmem97 | 3805 | 0.209 | -0.0813 | No |
| 7 | Trp53inp1 | 4457 | 0.174 | -0.1050 | No |
| 8 | Actg1 | 4724 | 0.161 | -0.1085 | No |
| 9 | Antxr2 | 5540 | 0.125 | -0.1445 | No |
| 10 | Ethe1 | 5578 | 0.124 | -0.1381 | No |
| 11 | Chka | 5630 | 0.122 | -0.1326 | No |
| 12 | Hmgcr | 6134 | 0.099 | -0.1533 | No |
| 13 | Plaur | 7381 | 0.050 | -0.2178 | No |
| 14 | Ctnnb1 | 7543 | 0.043 | -0.2236 | No |
| 15 | Stx5a | 7722 | 0.036 | -0.2309 | No |
| 16 | Anxa5 | 7749 | 0.035 | -0.2299 | No |
| 17 | Dhcr7 | 7758 | 0.035 | -0.2280 | No |
| 18 | Acss2 | 7837 | 0.032 | -0.2301 | No |
| 19 | Acat2 | 8062 | 0.023 | -0.2407 | No |
| 20 | Fads2 | 8426 | 0.009 | -0.2599 | No |
| 21 | Cyp51 | 8739 | -0.001 | -0.2768 | No |
| 22 | Nsdhl | 8851 | -0.007 | -0.2824 | No |
| 23 | Tnfrsf12a | 8856 | -0.007 | -0.2822 | No |
| 24 | Tm7sf2 | 8957 | -0.011 | -0.2869 | No |
| 25 | Abca2 | 9703 | -0.039 | -0.3248 | No |
| 26 | Gusb | 10221 | -0.060 | -0.3489 | No |
| 27 | Stard4 | 10246 | -0.061 | -0.3461 | No |
| 28 | Fasn | 10304 | -0.063 | -0.3450 | No |
| 29 | Atf5 | 10400 | -0.067 | -0.3456 | No |
| 30 | Errfi1 | 10512 | -0.071 | -0.3468 | No |
| 31 | Sqle | 10604 | -0.075 | -0.3467 | No |
| 32 | Srebf2 | 10768 | -0.082 | -0.3500 | No |
| 33 | Niban1 | 11094 | -0.095 | -0.3613 | No |
| 34 | Atxn2 | 11114 | -0.096 | -0.3558 | No |
| 35 | Lgals3 | 11255 | -0.101 | -0.3565 | No |
| 36 | Pmvk | 11465 | -0.110 | -0.3605 | No |
| 37 | Pparg | 11619 | -0.116 | -0.3609 | No |
| 38 | Fabp5 | 11697 | -0.119 | -0.3570 | No |
| 39 | Pdk3 | 11827 | -0.124 | -0.3556 | No |
| 40 | Aldoc | 11850 | -0.125 | -0.3483 | No |
| 41 | Plscr1 | 12113 | -0.137 | -0.3533 | No |
| 42 | Ebp | 13452 | -0.201 | -0.4125 | No |
| 43 | Alcam | 13964 | -0.229 | -0.4248 | No |
| 44 | Lss | 15386 | -0.314 | -0.4809 | Yes |
| 45 | S100a11 | 15715 | -0.337 | -0.4759 | Yes |
| 46 | Hmgcs1 | 15733 | -0.338 | -0.4539 | Yes |
| 47 | Pnrc1 | 16085 | -0.368 | -0.4480 | Yes |
| 48 | Trib3 | 16163 | -0.372 | -0.4269 | Yes |
| 49 | Mvk | 16388 | -0.394 | -0.4123 | Yes |
| 50 | Lgmn | 16404 | -0.395 | -0.3863 | Yes |
| 51 | Sc5d | 16807 | -0.434 | -0.3787 | Yes |
| 52 | Pcyt2 | 16975 | -0.454 | -0.3569 | Yes |
| 53 | Ech1 | 17042 | -0.463 | -0.3290 | Yes |
| 54 | Gnai1 | 17225 | -0.484 | -0.3061 | Yes |
| 55 | Ldlr | 17438 | -0.515 | -0.2826 | Yes |
| 56 | Hsd17b7 | 17541 | -0.533 | -0.2519 | Yes |
| 57 | Fdft1 | 17925 | -0.626 | -0.2303 | Yes |
| 58 | Cd9 | 18050 | -0.669 | -0.1916 | Yes |
| 59 | Gstm7 | 18097 | -0.692 | -0.1470 | Yes |
| 60 | Fdps | 18185 | -0.739 | -0.1015 | Yes |
| 61 | Fbxo6 | 18201 | -0.754 | -0.0510 | Yes |
| 62 | Mvd | 18344 | -0.916 | 0.0035 | Yes |