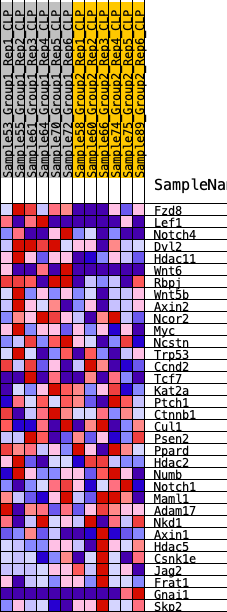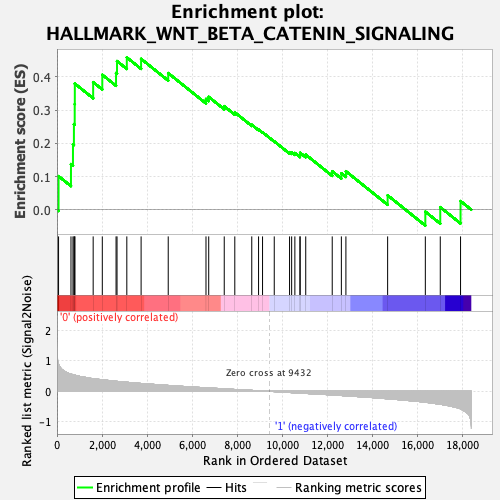
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group1_versus_Group2.Basophil_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_WNT_BETA_CATENIN_SIGNALING |
| Enrichment Score (ES) | 0.4582862 |
| Normalized Enrichment Score (NES) | 1.7087325 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.102700226 |
| FWER p-Value | 0.116 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Fzd8 | 69 | 0.896 | 0.1010 | Yes |
| 2 | Lef1 | 621 | 0.565 | 0.1372 | Yes |
| 3 | Notch4 | 710 | 0.548 | 0.1965 | Yes |
| 4 | Dvl2 | 752 | 0.539 | 0.2574 | Yes |
| 5 | Hdac11 | 787 | 0.531 | 0.3177 | Yes |
| 6 | Wnt6 | 792 | 0.531 | 0.3795 | Yes |
| 7 | Rbpj | 1607 | 0.414 | 0.3837 | Yes |
| 8 | Wnt5b | 2013 | 0.380 | 0.4061 | Yes |
| 9 | Axin2 | 2624 | 0.329 | 0.4113 | Yes |
| 10 | Ncor2 | 2667 | 0.326 | 0.4472 | Yes |
| 11 | Myc | 3102 | 0.297 | 0.4583 | Yes |
| 12 | Ncstn | 3737 | 0.258 | 0.4539 | No |
| 13 | Trp53 | 4942 | 0.193 | 0.4111 | No |
| 14 | Ccnd2 | 6620 | 0.115 | 0.3332 | No |
| 15 | Tcf7 | 6741 | 0.110 | 0.3395 | No |
| 16 | Kat2a | 7431 | 0.081 | 0.3115 | No |
| 17 | Ptch1 | 7900 | 0.060 | 0.2930 | No |
| 18 | Ctnnb1 | 8654 | 0.031 | 0.2556 | No |
| 19 | Cul1 | 8956 | 0.018 | 0.2414 | No |
| 20 | Psen2 | 9133 | 0.012 | 0.2331 | No |
| 21 | Ppard | 9653 | -0.007 | 0.2057 | No |
| 22 | Hdac2 | 10333 | -0.036 | 0.1730 | No |
| 23 | Numb | 10426 | -0.041 | 0.1728 | No |
| 24 | Notch1 | 10569 | -0.047 | 0.1706 | No |
| 25 | Maml1 | 10795 | -0.057 | 0.1650 | No |
| 26 | Adam17 | 10803 | -0.058 | 0.1714 | No |
| 27 | Nkd1 | 11052 | -0.068 | 0.1659 | No |
| 28 | Axin1 | 12229 | -0.118 | 0.1156 | No |
| 29 | Hdac5 | 12635 | -0.137 | 0.1096 | No |
| 30 | Csnk1e | 12837 | -0.146 | 0.1157 | No |
| 31 | Jag2 | 14691 | -0.243 | 0.0434 | No |
| 32 | Frat1 | 16366 | -0.358 | -0.0058 | No |
| 33 | Gnai1 | 17030 | -0.423 | 0.0077 | No |
| 34 | Skp2 | 17931 | -0.575 | 0.0260 | No |