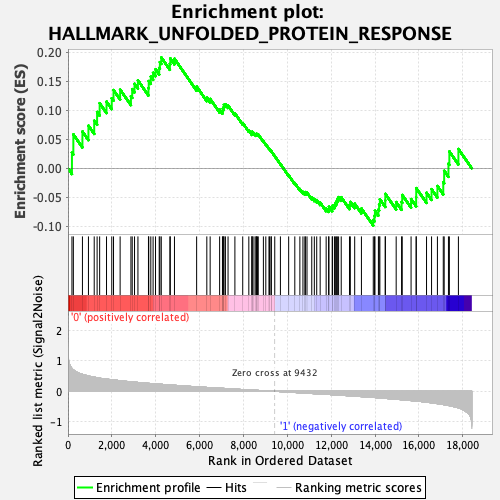
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group1_versus_Group2.Basophil_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_UNFOLDED_PROTEIN_RESPONSE |
| Enrichment Score (ES) | 0.19095202 |
| Normalized Enrichment Score (NES) | 0.8324145 |
| Nominal p-value | 0.6490486 |
| FDR q-value | 0.95434 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Nabp1 | 180 | 0.750 | 0.0272 | Yes |
| 2 | Edc4 | 244 | 0.705 | 0.0585 | Yes |
| 3 | Dnajb9 | 658 | 0.558 | 0.0635 | Yes |
| 4 | Exosc5 | 932 | 0.505 | 0.0735 | Yes |
| 5 | Gosr2 | 1195 | 0.465 | 0.0821 | Yes |
| 6 | Vegfa | 1327 | 0.448 | 0.0971 | Yes |
| 7 | Ero1a | 1443 | 0.433 | 0.1122 | Yes |
| 8 | Srprb | 1756 | 0.401 | 0.1149 | Yes |
| 9 | Asns | 1995 | 0.381 | 0.1207 | Yes |
| 10 | Bag3 | 2075 | 0.374 | 0.1348 | Yes |
| 11 | Atp6v0d1 | 2376 | 0.349 | 0.1356 | Yes |
| 12 | Herpud1 | 2869 | 0.313 | 0.1242 | Yes |
| 13 | Tspyl2 | 2927 | 0.309 | 0.1364 | Yes |
| 14 | Ddx10 | 3033 | 0.301 | 0.1455 | Yes |
| 15 | Nfya | 3188 | 0.292 | 0.1515 | Yes |
| 16 | Preb | 3668 | 0.261 | 0.1382 | Yes |
| 17 | H2ax | 3681 | 0.261 | 0.1504 | Yes |
| 18 | Fkbp14 | 3775 | 0.255 | 0.1579 | Yes |
| 19 | Ern1 | 3879 | 0.249 | 0.1646 | Yes |
| 20 | Eif4ebp1 | 3990 | 0.243 | 0.1706 | Yes |
| 21 | Slc7a5 | 4158 | 0.235 | 0.1730 | Yes |
| 22 | Sec11a | 4189 | 0.234 | 0.1830 | Yes |
| 23 | Sdad1 | 4252 | 0.231 | 0.1910 | Yes |
| 24 | Srpr | 4645 | 0.210 | 0.1799 | No |
| 25 | Arfgap1 | 4663 | 0.209 | 0.1893 | No |
| 26 | Slc30a5 | 4853 | 0.198 | 0.1887 | No |
| 27 | Eef2 | 5864 | 0.148 | 0.1408 | No |
| 28 | Nop56 | 6328 | 0.127 | 0.1218 | No |
| 29 | Aldh18a1 | 6480 | 0.121 | 0.1195 | No |
| 30 | Eif4a2 | 6912 | 0.102 | 0.1010 | No |
| 31 | Atf3 | 7041 | 0.097 | 0.0989 | No |
| 32 | Rps14 | 7060 | 0.096 | 0.1026 | No |
| 33 | Pdia5 | 7098 | 0.095 | 0.1053 | No |
| 34 | Paip1 | 7108 | 0.095 | 0.1095 | No |
| 35 | Dkc1 | 7176 | 0.092 | 0.1104 | No |
| 36 | Cks1b | 7296 | 0.086 | 0.1081 | No |
| 37 | Zbtb17 | 7608 | 0.072 | 0.0947 | No |
| 38 | Xpot | 7965 | 0.057 | 0.0781 | No |
| 39 | Mtrex | 8243 | 0.046 | 0.0652 | No |
| 40 | Eif4e | 8386 | 0.041 | 0.0595 | No |
| 41 | Hyou1 | 8389 | 0.041 | 0.0614 | No |
| 42 | Sec31a | 8393 | 0.041 | 0.0632 | No |
| 43 | Nfyb | 8477 | 0.038 | 0.0606 | No |
| 44 | Imp3 | 8564 | 0.035 | 0.0576 | No |
| 45 | Exosc1 | 8582 | 0.034 | 0.0584 | No |
| 46 | Slc1a4 | 8590 | 0.034 | 0.0597 | No |
| 47 | Exosc4 | 8640 | 0.031 | 0.0586 | No |
| 48 | Nolc1 | 8672 | 0.030 | 0.0584 | No |
| 49 | Lsm4 | 8907 | 0.020 | 0.0466 | No |
| 50 | Ywhaz | 9017 | 0.016 | 0.0414 | No |
| 51 | Parn | 9162 | 0.010 | 0.0340 | No |
| 52 | Atf6 | 9190 | 0.009 | 0.0330 | No |
| 53 | Ddit4 | 9257 | 0.007 | 0.0298 | No |
| 54 | Yif1a | 9268 | 0.006 | 0.0295 | No |
| 55 | Xbp1 | 9425 | 0.000 | 0.0210 | No |
| 56 | Khsrp | 9683 | -0.008 | 0.0074 | No |
| 57 | Wipi1 | 10059 | -0.024 | -0.0119 | No |
| 58 | Kif5b | 10339 | -0.036 | -0.0254 | No |
| 59 | Lsm1 | 10570 | -0.047 | -0.0356 | No |
| 60 | Cnot4 | 10710 | -0.054 | -0.0406 | No |
| 61 | Tatdn2 | 10799 | -0.057 | -0.0425 | No |
| 62 | Cnot2 | 10821 | -0.059 | -0.0408 | No |
| 63 | Npm1 | 10895 | -0.062 | -0.0417 | No |
| 64 | Fus | 11112 | -0.070 | -0.0500 | No |
| 65 | Gemin4 | 11229 | -0.075 | -0.0527 | No |
| 66 | Cxxc1 | 11338 | -0.080 | -0.0546 | No |
| 67 | Dctn1 | 11495 | -0.087 | -0.0589 | No |
| 68 | Rrp9 | 11768 | -0.096 | -0.0690 | No |
| 69 | Arxes2 | 11879 | -0.102 | -0.0700 | No |
| 70 | Nop14 | 11896 | -0.102 | -0.0658 | No |
| 71 | Eif2s1 | 12043 | -0.109 | -0.0684 | No |
| 72 | Eif4g1 | 12061 | -0.110 | -0.0639 | No |
| 73 | Dnajc3 | 12151 | -0.114 | -0.0631 | No |
| 74 | Mthfd2 | 12188 | -0.116 | -0.0594 | No |
| 75 | Banf1 | 12244 | -0.118 | -0.0565 | No |
| 76 | Eif4a3 | 12279 | -0.120 | -0.0525 | No |
| 77 | Cnot6 | 12335 | -0.123 | -0.0494 | No |
| 78 | Nhp2 | 12455 | -0.128 | -0.0496 | No |
| 79 | Exosc10 | 12832 | -0.146 | -0.0630 | No |
| 80 | Atf4 | 12870 | -0.147 | -0.0577 | No |
| 81 | Spcs1 | 13069 | -0.157 | -0.0608 | No |
| 82 | Dnaja4 | 13376 | -0.172 | -0.0690 | No |
| 83 | Serp1 | 13916 | -0.199 | -0.0886 | No |
| 84 | Psat1 | 13969 | -0.202 | -0.0815 | No |
| 85 | Shc1 | 13993 | -0.203 | -0.0728 | No |
| 86 | Edem1 | 14150 | -0.212 | -0.0708 | No |
| 87 | Hsp90b1 | 14183 | -0.214 | -0.0620 | No |
| 88 | Pdia6 | 14217 | -0.216 | -0.0532 | No |
| 89 | Ssr1 | 14463 | -0.231 | -0.0552 | No |
| 90 | Exoc2 | 14466 | -0.231 | -0.0439 | No |
| 91 | Eif2ak3 | 14960 | -0.260 | -0.0580 | No |
| 92 | Tubb2a | 15206 | -0.274 | -0.0578 | No |
| 93 | Exosc2 | 15236 | -0.276 | -0.0458 | No |
| 94 | Eif4a1 | 15639 | -0.303 | -0.0528 | No |
| 95 | Calr | 15871 | -0.320 | -0.0496 | No |
| 96 | Dcp2 | 15875 | -0.320 | -0.0340 | No |
| 97 | Dcp1a | 16344 | -0.357 | -0.0419 | No |
| 98 | Exosc9 | 16573 | -0.378 | -0.0357 | No |
| 99 | Wfs1 | 16840 | -0.404 | -0.0303 | No |
| 100 | Hspa5 | 17107 | -0.432 | -0.0235 | No |
| 101 | Cebpb | 17148 | -0.439 | -0.0041 | No |
| 102 | Hspa9 | 17343 | -0.464 | 0.0083 | No |
| 103 | Pop4 | 17385 | -0.469 | 0.0292 | No |
| 104 | Cebpg | 17797 | -0.540 | 0.0334 | No |

