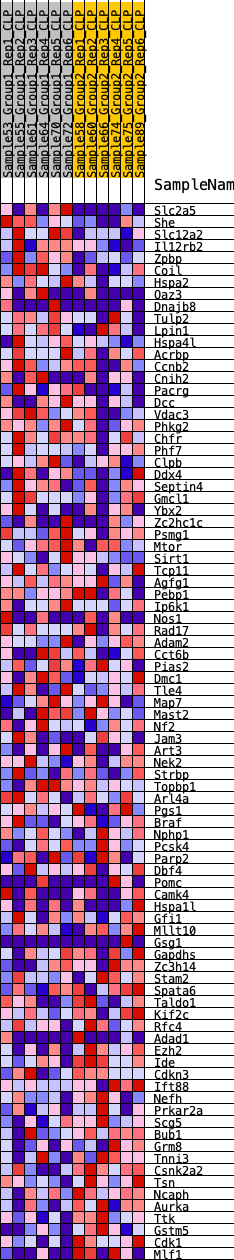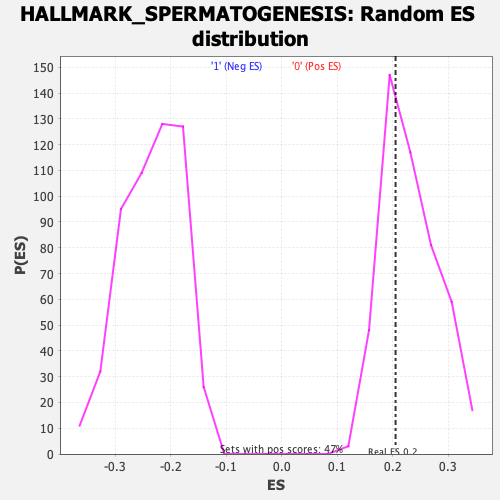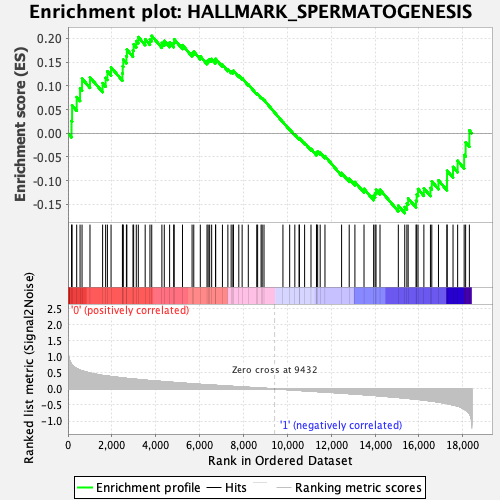
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group1_versus_Group2.Basophil_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_SPERMATOGENESIS |
| Enrichment Score (ES) | 0.20532504 |
| Normalized Enrichment Score (NES) | 0.8840974 |
| Nominal p-value | 0.62711865 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Slc2a5 | 160 | 0.766 | 0.0263 | Yes |
| 2 | She | 187 | 0.744 | 0.0589 | Yes |
| 3 | Slc12a2 | 396 | 0.628 | 0.0762 | Yes |
| 4 | Il12rb2 | 550 | 0.581 | 0.0944 | Yes |
| 5 | Zpbp | 640 | 0.562 | 0.1153 | Yes |
| 6 | Coil | 1002 | 0.491 | 0.1180 | Yes |
| 7 | Hspa2 | 1578 | 0.416 | 0.1057 | Yes |
| 8 | Oaz3 | 1715 | 0.405 | 0.1167 | Yes |
| 9 | Dnajb8 | 1796 | 0.397 | 0.1305 | Yes |
| 10 | Tulp2 | 1963 | 0.384 | 0.1390 | Yes |
| 11 | Lpin1 | 2481 | 0.341 | 0.1264 | Yes |
| 12 | Hspa4l | 2494 | 0.340 | 0.1413 | Yes |
| 13 | Acrbp | 2519 | 0.338 | 0.1554 | Yes |
| 14 | Ccnb2 | 2663 | 0.326 | 0.1625 | Yes |
| 15 | Cnih2 | 2683 | 0.325 | 0.1763 | Yes |
| 16 | Pacrg | 2962 | 0.307 | 0.1752 | Yes |
| 17 | Dcc | 2992 | 0.304 | 0.1875 | Yes |
| 18 | Vdac3 | 3113 | 0.296 | 0.1945 | Yes |
| 19 | Phkg2 | 3205 | 0.291 | 0.2028 | Yes |
| 20 | Chfr | 3518 | 0.271 | 0.1982 | Yes |
| 21 | Phf7 | 3736 | 0.258 | 0.1981 | Yes |
| 22 | Clpb | 3817 | 0.253 | 0.2053 | Yes |
| 23 | Ddx4 | 4280 | 0.229 | 0.1906 | No |
| 24 | Septin4 | 4389 | 0.223 | 0.1949 | No |
| 25 | Gmcl1 | 4632 | 0.211 | 0.1913 | No |
| 26 | Ybx2 | 4818 | 0.200 | 0.1903 | No |
| 27 | Zc2hc1c | 4840 | 0.199 | 0.1983 | No |
| 28 | Psmg1 | 5219 | 0.179 | 0.1858 | No |
| 29 | Mtor | 5656 | 0.157 | 0.1692 | No |
| 30 | Sirt1 | 5733 | 0.154 | 0.1721 | No |
| 31 | Tcp11 | 6031 | 0.139 | 0.1623 | No |
| 32 | Agfg1 | 6337 | 0.127 | 0.1514 | No |
| 33 | Pebp1 | 6389 | 0.125 | 0.1543 | No |
| 34 | Ip6k1 | 6458 | 0.122 | 0.1562 | No |
| 35 | Nos1 | 6549 | 0.118 | 0.1566 | No |
| 36 | Rad17 | 6724 | 0.110 | 0.1522 | No |
| 37 | Adam2 | 6733 | 0.110 | 0.1568 | No |
| 38 | Cct6b | 7040 | 0.097 | 0.1445 | No |
| 39 | Pias2 | 7288 | 0.086 | 0.1350 | No |
| 40 | Dmc1 | 7437 | 0.080 | 0.1306 | No |
| 41 | Tle4 | 7517 | 0.077 | 0.1298 | No |
| 42 | Map7 | 7541 | 0.075 | 0.1320 | No |
| 43 | Mast2 | 7786 | 0.065 | 0.1216 | No |
| 44 | Nf2 | 7938 | 0.058 | 0.1161 | No |
| 45 | Jam3 | 8219 | 0.047 | 0.1029 | No |
| 46 | Art3 | 8604 | 0.033 | 0.0835 | No |
| 47 | Nek2 | 8644 | 0.031 | 0.0828 | No |
| 48 | Strbp | 8802 | 0.024 | 0.0753 | No |
| 49 | Topbp1 | 8851 | 0.022 | 0.0737 | No |
| 50 | Arl4a | 8935 | 0.019 | 0.0701 | No |
| 51 | Pgs1 | 9798 | -0.013 | 0.0236 | No |
| 52 | Braf | 10105 | -0.026 | 0.0081 | No |
| 53 | Nphp1 | 10340 | -0.037 | -0.0030 | No |
| 54 | Pcsk4 | 10533 | -0.045 | -0.0114 | No |
| 55 | Parp2 | 10550 | -0.046 | -0.0101 | No |
| 56 | Dbf4 | 10786 | -0.057 | -0.0204 | No |
| 57 | Pomc | 11077 | -0.069 | -0.0330 | No |
| 58 | Camk4 | 11327 | -0.080 | -0.0430 | No |
| 59 | Hspa1l | 11337 | -0.080 | -0.0398 | No |
| 60 | Gfi1 | 11381 | -0.082 | -0.0384 | No |
| 61 | Mllt10 | 11496 | -0.087 | -0.0407 | No |
| 62 | Gsg1 | 11714 | -0.095 | -0.0482 | No |
| 63 | Gapdhs | 12472 | -0.129 | -0.0836 | No |
| 64 | Zc3h14 | 12819 | -0.145 | -0.0959 | No |
| 65 | Stam2 | 13079 | -0.158 | -0.1028 | No |
| 66 | Spata6 | 13494 | -0.178 | -0.1173 | No |
| 67 | Taldo1 | 13932 | -0.200 | -0.1320 | No |
| 68 | Kif2c | 13998 | -0.204 | -0.1262 | No |
| 69 | Rfc4 | 14041 | -0.206 | -0.1191 | No |
| 70 | Adad1 | 14223 | -0.217 | -0.1191 | No |
| 71 | Ezh2 | 15058 | -0.265 | -0.1525 | No |
| 72 | Ide | 15351 | -0.282 | -0.1555 | No |
| 73 | Cdkn3 | 15448 | -0.288 | -0.1476 | No |
| 74 | Ift88 | 15506 | -0.294 | -0.1373 | No |
| 75 | Nefh | 15865 | -0.319 | -0.1422 | No |
| 76 | Prkar2a | 15896 | -0.322 | -0.1292 | No |
| 77 | Scg5 | 15957 | -0.326 | -0.1175 | No |
| 78 | Bub1 | 16223 | -0.347 | -0.1162 | No |
| 79 | Grm8 | 16522 | -0.372 | -0.1154 | No |
| 80 | Tnni3 | 16584 | -0.379 | -0.1014 | No |
| 81 | Csnk2a2 | 16888 | -0.409 | -0.0993 | No |
| 82 | Tsn | 17275 | -0.455 | -0.0995 | No |
| 83 | Ncaph | 17281 | -0.455 | -0.0790 | No |
| 84 | Aurka | 17552 | -0.497 | -0.0710 | No |
| 85 | Ttk | 17762 | -0.532 | -0.0581 | No |
| 86 | Gstm5 | 18059 | -0.624 | -0.0457 | No |
| 87 | Cdk1 | 18126 | -0.659 | -0.0192 | No |
| 88 | Mlf1 | 18297 | -0.756 | 0.0061 | No |