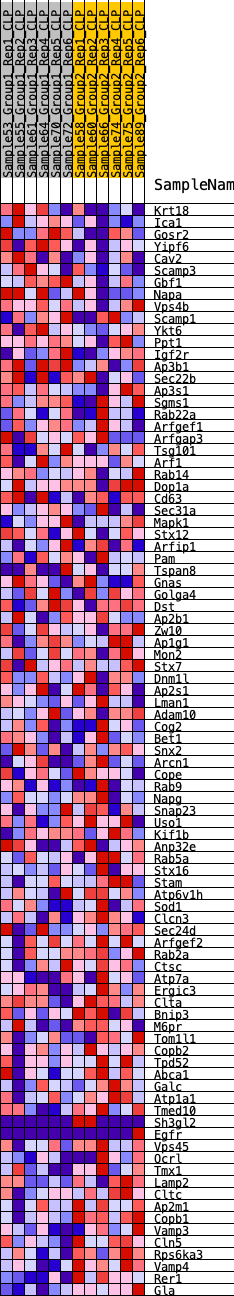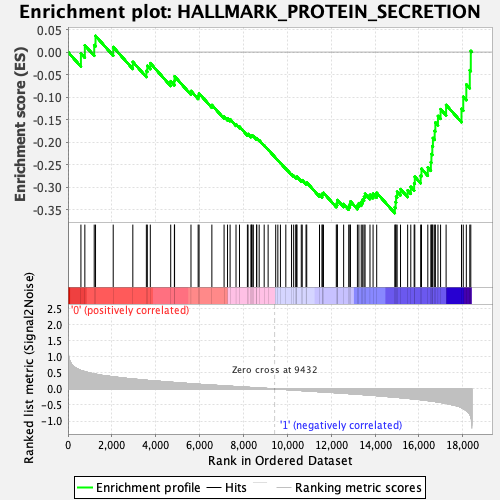
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group1_versus_Group2.Basophil_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_PROTEIN_SECRETION |
| Enrichment Score (ES) | -0.3572707 |
| Normalized Enrichment Score (NES) | -1.5327617 |
| Nominal p-value | 0.04032258 |
| FDR q-value | 0.13105033 |
| FWER p-Value | 0.314 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Krt18 | 589 | 0.571 | -0.0028 | No |
| 2 | Ica1 | 768 | 0.535 | 0.0149 | No |
| 3 | Gosr2 | 1195 | 0.465 | 0.0156 | No |
| 4 | Yipf6 | 1255 | 0.458 | 0.0359 | No |
| 5 | Cav2 | 2061 | 0.375 | 0.0112 | No |
| 6 | Scamp3 | 2955 | 0.307 | -0.0218 | No |
| 7 | Gbf1 | 3572 | 0.267 | -0.0417 | No |
| 8 | Napa | 3616 | 0.264 | -0.0304 | No |
| 9 | Vps4b | 3752 | 0.257 | -0.0246 | No |
| 10 | Scamp1 | 4682 | 0.208 | -0.0647 | No |
| 11 | Ykt6 | 4850 | 0.198 | -0.0636 | No |
| 12 | Ppt1 | 4858 | 0.198 | -0.0538 | No |
| 13 | Igf2r | 5606 | 0.160 | -0.0863 | No |
| 14 | Ap3b1 | 5933 | 0.144 | -0.0967 | No |
| 15 | Sec22b | 5975 | 0.142 | -0.0917 | No |
| 16 | Ap3s1 | 6558 | 0.118 | -0.1174 | No |
| 17 | Sgms1 | 7115 | 0.095 | -0.1429 | No |
| 18 | Rab22a | 7272 | 0.087 | -0.1470 | No |
| 19 | Arfgef1 | 7389 | 0.082 | -0.1491 | No |
| 20 | Arfgap3 | 7659 | 0.070 | -0.1601 | No |
| 21 | Tsg101 | 7818 | 0.064 | -0.1655 | No |
| 22 | Arf1 | 8178 | 0.049 | -0.1826 | No |
| 23 | Rab14 | 8207 | 0.047 | -0.1817 | No |
| 24 | Dop1a | 8339 | 0.043 | -0.1866 | No |
| 25 | Cd63 | 8366 | 0.042 | -0.1859 | No |
| 26 | Sec31a | 8393 | 0.041 | -0.1852 | No |
| 27 | Mapk1 | 8448 | 0.040 | -0.1861 | No |
| 28 | Stx12 | 8599 | 0.034 | -0.1926 | No |
| 29 | Arfip1 | 8602 | 0.034 | -0.1910 | No |
| 30 | Pam | 8715 | 0.028 | -0.1957 | No |
| 31 | Tspan8 | 8943 | 0.019 | -0.2071 | No |
| 32 | Gnas | 9123 | 0.012 | -0.2162 | No |
| 33 | Golga4 | 9468 | -0.001 | -0.2350 | No |
| 34 | Dst | 9566 | -0.004 | -0.2401 | No |
| 35 | Ap2b1 | 9684 | -0.008 | -0.2460 | No |
| 36 | Zw10 | 9929 | -0.018 | -0.2584 | No |
| 37 | Ap1g1 | 10190 | -0.030 | -0.2711 | No |
| 38 | Mon2 | 10287 | -0.034 | -0.2746 | No |
| 39 | Stx7 | 10380 | -0.038 | -0.2776 | No |
| 40 | Dnm1l | 10424 | -0.041 | -0.2779 | No |
| 41 | Ap2s1 | 10435 | -0.041 | -0.2763 | No |
| 42 | Lman1 | 10634 | -0.050 | -0.2846 | No |
| 43 | Adam10 | 10677 | -0.052 | -0.2842 | No |
| 44 | Cog2 | 10851 | -0.060 | -0.2906 | No |
| 45 | Bet1 | 10883 | -0.062 | -0.2891 | No |
| 46 | Snx2 | 11465 | -0.085 | -0.3164 | No |
| 47 | Arcn1 | 11582 | -0.090 | -0.3182 | No |
| 48 | Cope | 11591 | -0.090 | -0.3140 | No |
| 49 | Rab9 | 11646 | -0.093 | -0.3122 | No |
| 50 | Napg | 12230 | -0.118 | -0.3380 | No |
| 51 | Snap23 | 12269 | -0.119 | -0.3339 | No |
| 52 | Uso1 | 12278 | -0.120 | -0.3282 | No |
| 53 | Kif1b | 12565 | -0.133 | -0.3370 | No |
| 54 | Anp32e | 12797 | -0.144 | -0.3422 | No |
| 55 | Rab5a | 12848 | -0.147 | -0.3374 | No |
| 56 | Stx16 | 12876 | -0.148 | -0.3313 | No |
| 57 | Stam | 13193 | -0.163 | -0.3401 | No |
| 58 | Atp6v1h | 13260 | -0.166 | -0.3352 | No |
| 59 | Sod1 | 13368 | -0.171 | -0.3322 | No |
| 60 | Clcn3 | 13435 | -0.175 | -0.3269 | No |
| 61 | Sec24d | 13495 | -0.178 | -0.3209 | No |
| 62 | Arfgef2 | 13542 | -0.180 | -0.3142 | No |
| 63 | Rab2a | 13766 | -0.192 | -0.3165 | No |
| 64 | Ctsc | 13907 | -0.199 | -0.3139 | No |
| 65 | Atp7a | 14072 | -0.208 | -0.3122 | No |
| 66 | Ergic3 | 14898 | -0.257 | -0.3441 | Yes |
| 67 | Clta | 14935 | -0.259 | -0.3328 | Yes |
| 68 | Bnip3 | 14957 | -0.260 | -0.3205 | Yes |
| 69 | M6pr | 15003 | -0.263 | -0.3095 | Yes |
| 70 | Tom1l1 | 15158 | -0.271 | -0.3040 | Yes |
| 71 | Copb2 | 15483 | -0.292 | -0.3067 | Yes |
| 72 | Tpd52 | 15626 | -0.303 | -0.2989 | Yes |
| 73 | Abca1 | 15780 | -0.313 | -0.2912 | Yes |
| 74 | Galc | 15809 | -0.315 | -0.2765 | Yes |
| 75 | Atp1a1 | 16077 | -0.333 | -0.2740 | Yes |
| 76 | Tmed10 | 16114 | -0.336 | -0.2587 | Yes |
| 77 | Sh3gl2 | 16402 | -0.361 | -0.2558 | Yes |
| 78 | Egfr | 16542 | -0.374 | -0.2442 | Yes |
| 79 | Vps45 | 16566 | -0.377 | -0.2261 | Yes |
| 80 | Ocrl | 16608 | -0.381 | -0.2088 | Yes |
| 81 | Tmx1 | 16635 | -0.383 | -0.1905 | Yes |
| 82 | Lamp2 | 16715 | -0.390 | -0.1748 | Yes |
| 83 | Cltc | 16747 | -0.394 | -0.1562 | Yes |
| 84 | Ap2m1 | 16865 | -0.407 | -0.1417 | Yes |
| 85 | Copb1 | 16984 | -0.418 | -0.1267 | Yes |
| 86 | Vamp3 | 17236 | -0.449 | -0.1173 | Yes |
| 87 | Cln5 | 17938 | -0.577 | -0.1259 | Yes |
| 88 | Rps6ka3 | 18018 | -0.608 | -0.0990 | Yes |
| 89 | Vamp4 | 18158 | -0.675 | -0.0720 | Yes |
| 90 | Rer1 | 18316 | -0.784 | -0.0403 | Yes |
| 91 | Gla | 18362 | -0.881 | 0.0025 | Yes |