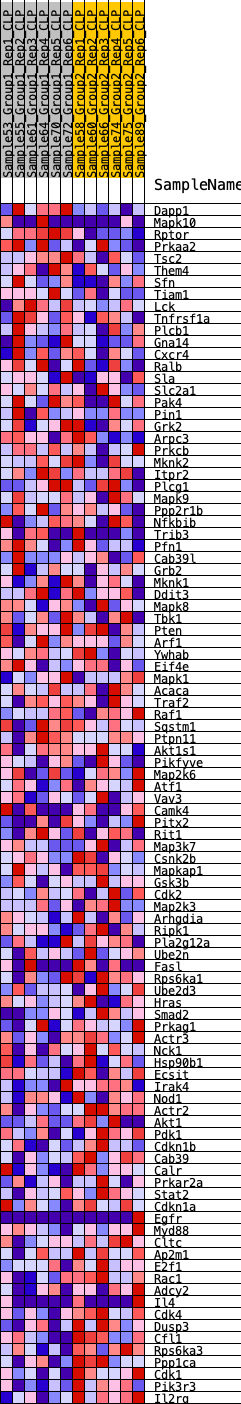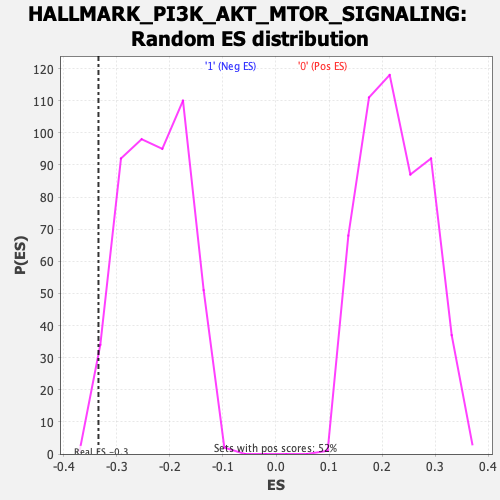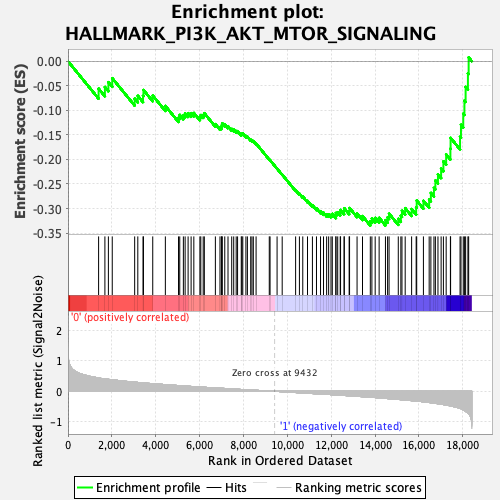
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group1_versus_Group2.Basophil_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_PI3K_AKT_MTOR_SIGNALING |
| Enrichment Score (ES) | -0.33483937 |
| Normalized Enrichment Score (NES) | -1.4665383 |
| Nominal p-value | 0.010351967 |
| FDR q-value | 0.15940455 |
| FWER p-Value | 0.444 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Dapp1 | 1395 | 0.438 | -0.0558 | No |
| 2 | Mapk10 | 1682 | 0.408 | -0.0524 | No |
| 3 | Rptor | 1841 | 0.394 | -0.0426 | No |
| 4 | Prkaa2 | 2017 | 0.379 | -0.0345 | No |
| 5 | Tsc2 | 3039 | 0.301 | -0.0762 | No |
| 6 | Them4 | 3182 | 0.292 | -0.0704 | No |
| 7 | Sfn | 3420 | 0.277 | -0.0704 | No |
| 8 | Tiam1 | 3440 | 0.276 | -0.0585 | No |
| 9 | Lck | 3863 | 0.250 | -0.0699 | No |
| 10 | Tnfrsf1a | 4438 | 0.220 | -0.0910 | No |
| 11 | Plcb1 | 5038 | 0.188 | -0.1150 | No |
| 12 | Gna14 | 5094 | 0.186 | -0.1093 | No |
| 13 | Cxcr4 | 5259 | 0.177 | -0.1100 | No |
| 14 | Ralb | 5339 | 0.173 | -0.1062 | No |
| 15 | Sla | 5475 | 0.167 | -0.1058 | No |
| 16 | Slc2a1 | 5612 | 0.160 | -0.1058 | No |
| 17 | Pak4 | 5736 | 0.154 | -0.1053 | No |
| 18 | Pin1 | 6012 | 0.140 | -0.1138 | No |
| 19 | Grk2 | 6054 | 0.138 | -0.1096 | No |
| 20 | Arpc3 | 6163 | 0.134 | -0.1092 | No |
| 21 | Prkcb | 6215 | 0.132 | -0.1058 | No |
| 22 | Mknk2 | 6718 | 0.111 | -0.1281 | No |
| 23 | Itpr2 | 6928 | 0.102 | -0.1348 | No |
| 24 | Plcg1 | 6994 | 0.099 | -0.1337 | No |
| 25 | Mapk9 | 7014 | 0.098 | -0.1301 | No |
| 26 | Ppp2r1b | 7030 | 0.098 | -0.1264 | No |
| 27 | Nfkbib | 7146 | 0.093 | -0.1283 | No |
| 28 | Trib3 | 7294 | 0.086 | -0.1324 | No |
| 29 | Pfn1 | 7463 | 0.079 | -0.1379 | No |
| 30 | Cab39l | 7554 | 0.074 | -0.1393 | No |
| 31 | Grb2 | 7667 | 0.070 | -0.1422 | No |
| 32 | Mknk1 | 7730 | 0.067 | -0.1424 | No |
| 33 | Ddit3 | 7898 | 0.060 | -0.1487 | No |
| 34 | Mapk8 | 7926 | 0.059 | -0.1475 | No |
| 35 | Tbk1 | 7968 | 0.057 | -0.1470 | No |
| 36 | Pten | 8110 | 0.052 | -0.1523 | No |
| 37 | Arf1 | 8178 | 0.049 | -0.1537 | No |
| 38 | Ywhab | 8329 | 0.043 | -0.1599 | No |
| 39 | Eif4e | 8386 | 0.041 | -0.1610 | No |
| 40 | Mapk1 | 8448 | 0.040 | -0.1625 | No |
| 41 | Acaca | 8574 | 0.035 | -0.1677 | No |
| 42 | Traf2 | 9175 | 0.010 | -0.2000 | No |
| 43 | Raf1 | 9211 | 0.008 | -0.2015 | No |
| 44 | Sqstm1 | 9531 | -0.003 | -0.2188 | No |
| 45 | Ptpn11 | 9764 | -0.012 | -0.2310 | No |
| 46 | Akt1s1 | 10376 | -0.038 | -0.2626 | No |
| 47 | Pikfyve | 10549 | -0.046 | -0.2698 | No |
| 48 | Map2k6 | 10702 | -0.053 | -0.2756 | No |
| 49 | Atf1 | 10918 | -0.063 | -0.2844 | No |
| 50 | Vav3 | 11142 | -0.071 | -0.2933 | No |
| 51 | Camk4 | 11327 | -0.080 | -0.2996 | No |
| 52 | Pitx2 | 11512 | -0.087 | -0.3056 | No |
| 53 | Rit1 | 11648 | -0.093 | -0.3086 | No |
| 54 | Map3k7 | 11790 | -0.097 | -0.3118 | No |
| 55 | Csnk2b | 11882 | -0.102 | -0.3121 | No |
| 56 | Mapkap1 | 11989 | -0.107 | -0.3129 | No |
| 57 | Gsk3b | 12048 | -0.110 | -0.3109 | No |
| 58 | Cdk2 | 12212 | -0.117 | -0.3144 | No |
| 59 | Map2k3 | 12213 | -0.117 | -0.3090 | No |
| 60 | Arhgdia | 12295 | -0.121 | -0.3077 | No |
| 61 | Ripk1 | 12409 | -0.126 | -0.3080 | No |
| 62 | Pla2g12a | 12425 | -0.127 | -0.3029 | No |
| 63 | Ube2n | 12586 | -0.134 | -0.3054 | No |
| 64 | Fasl | 12594 | -0.134 | -0.2996 | No |
| 65 | Rps6ka1 | 12816 | -0.145 | -0.3049 | No |
| 66 | Ube2d3 | 12835 | -0.146 | -0.2990 | No |
| 67 | Hras | 13177 | -0.162 | -0.3101 | No |
| 68 | Smad2 | 13425 | -0.174 | -0.3154 | No |
| 69 | Prkag1 | 13781 | -0.193 | -0.3258 | Yes |
| 70 | Actr3 | 13853 | -0.197 | -0.3205 | Yes |
| 71 | Nck1 | 14003 | -0.204 | -0.3192 | Yes |
| 72 | Hsp90b1 | 14183 | -0.214 | -0.3190 | Yes |
| 73 | Ecsit | 14471 | -0.231 | -0.3239 | Yes |
| 74 | Irak4 | 14569 | -0.237 | -0.3181 | Yes |
| 75 | Nod1 | 14636 | -0.240 | -0.3105 | Yes |
| 76 | Actr2 | 15056 | -0.265 | -0.3211 | Yes |
| 77 | Akt1 | 15173 | -0.272 | -0.3147 | Yes |
| 78 | Pdk1 | 15224 | -0.275 | -0.3046 | Yes |
| 79 | Cdkn1b | 15371 | -0.283 | -0.2994 | Yes |
| 80 | Cab39 | 15664 | -0.305 | -0.3011 | Yes |
| 81 | Calr | 15871 | -0.320 | -0.2974 | Yes |
| 82 | Prkar2a | 15896 | -0.322 | -0.2837 | Yes |
| 83 | Stat2 | 16202 | -0.344 | -0.2843 | Yes |
| 84 | Cdkn1a | 16464 | -0.366 | -0.2815 | Yes |
| 85 | Egfr | 16542 | -0.374 | -0.2682 | Yes |
| 86 | Myd88 | 16682 | -0.387 | -0.2578 | Yes |
| 87 | Cltc | 16747 | -0.394 | -0.2429 | Yes |
| 88 | Ap2m1 | 16865 | -0.407 | -0.2303 | Yes |
| 89 | E2f1 | 17009 | -0.421 | -0.2185 | Yes |
| 90 | Rac1 | 17113 | -0.433 | -0.2039 | Yes |
| 91 | Adcy2 | 17239 | -0.450 | -0.1898 | Yes |
| 92 | Il4 | 17432 | -0.477 | -0.1780 | Yes |
| 93 | Cdk4 | 17441 | -0.478 | -0.1561 | Yes |
| 94 | Dusp3 | 17872 | -0.558 | -0.1536 | Yes |
| 95 | Cfl1 | 17914 | -0.568 | -0.1293 | Yes |
| 96 | Rps6ka3 | 18018 | -0.608 | -0.1066 | Yes |
| 97 | Ppp1ca | 18071 | -0.631 | -0.0800 | Yes |
| 98 | Cdk1 | 18126 | -0.659 | -0.0523 | Yes |
| 99 | Pik3r3 | 18231 | -0.713 | -0.0247 | Yes |
| 100 | Il2rg | 18264 | -0.736 | 0.0079 | Yes |