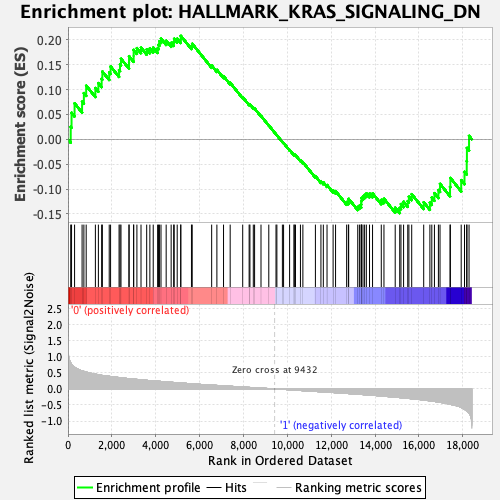
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group1_versus_Group2.Basophil_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_KRAS_SIGNALING_DN |
| Enrichment Score (ES) | 0.20776261 |
| Normalized Enrichment Score (NES) | 0.84027475 |
| Nominal p-value | 0.84250474 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Atp4a | 124 | 0.810 | 0.0251 | Yes |
| 2 | Brdt | 162 | 0.766 | 0.0533 | Yes |
| 3 | Zbtb16 | 300 | 0.671 | 0.0722 | Yes |
| 4 | Prodh | 642 | 0.562 | 0.0757 | Yes |
| 5 | Kmt2d | 724 | 0.545 | 0.0928 | Yes |
| 6 | Btg2 | 829 | 0.524 | 0.1077 | Yes |
| 7 | Sptbn2 | 1248 | 0.459 | 0.1030 | Yes |
| 8 | Nr4a2 | 1385 | 0.439 | 0.1129 | Yes |
| 9 | Kcnn1 | 1534 | 0.422 | 0.1214 | Yes |
| 10 | Kcnd1 | 1566 | 0.417 | 0.1361 | Yes |
| 11 | Pdk2 | 1879 | 0.391 | 0.1345 | Yes |
| 12 | Nr6a1 | 1941 | 0.385 | 0.1463 | Yes |
| 13 | Chst2 | 2328 | 0.352 | 0.1391 | Yes |
| 14 | Ngb | 2372 | 0.349 | 0.1505 | Yes |
| 15 | Htr1d | 2415 | 0.346 | 0.1618 | Yes |
| 16 | Edar | 2781 | 0.319 | 0.1544 | Yes |
| 17 | Prkn | 2783 | 0.319 | 0.1669 | Yes |
| 18 | Dcc | 2992 | 0.304 | 0.1676 | Yes |
| 19 | Snn | 2993 | 0.304 | 0.1795 | Yes |
| 20 | Itgb1bp2 | 3141 | 0.295 | 0.1831 | Yes |
| 21 | Arpp21 | 3328 | 0.284 | 0.1842 | Yes |
| 22 | Mfsd6 | 3587 | 0.266 | 0.1806 | Yes |
| 23 | Ypel1 | 3732 | 0.258 | 0.1829 | Yes |
| 24 | Thnsl2 | 3885 | 0.249 | 0.1843 | Yes |
| 25 | Mast3 | 4086 | 0.238 | 0.1828 | Yes |
| 26 | Slc25a23 | 4128 | 0.237 | 0.1899 | Yes |
| 27 | Cd80 | 4168 | 0.235 | 0.1970 | Yes |
| 28 | Zfp112 | 4239 | 0.231 | 0.2023 | Yes |
| 29 | Magix | 4475 | 0.218 | 0.1980 | Yes |
| 30 | Plag1 | 4702 | 0.206 | 0.1938 | Yes |
| 31 | Ybx2 | 4818 | 0.200 | 0.1954 | Yes |
| 32 | Zc2hc1c | 4840 | 0.199 | 0.2021 | Yes |
| 33 | Msh5 | 4978 | 0.192 | 0.2021 | Yes |
| 34 | Asb7 | 5139 | 0.184 | 0.2006 | Yes |
| 35 | Egf | 5142 | 0.184 | 0.2078 | Yes |
| 36 | Abcg4 | 5628 | 0.159 | 0.1875 | No |
| 37 | Thrb | 5662 | 0.157 | 0.1919 | No |
| 38 | Nos1 | 6549 | 0.118 | 0.1482 | No |
| 39 | Ryr1 | 6787 | 0.107 | 0.1395 | No |
| 40 | Stag3 | 7090 | 0.096 | 0.1267 | No |
| 41 | Dtnb | 7393 | 0.082 | 0.1135 | No |
| 42 | Pdcd1 | 7964 | 0.057 | 0.0846 | No |
| 43 | Slc16a7 | 8257 | 0.046 | 0.0705 | No |
| 44 | Gp1ba | 8293 | 0.045 | 0.0703 | No |
| 45 | Sidt1 | 8447 | 0.040 | 0.0635 | No |
| 46 | Klk8 | 8506 | 0.037 | 0.0618 | No |
| 47 | Tfcp2l1 | 8798 | 0.024 | 0.0469 | No |
| 48 | Arhgdig | 9153 | 0.011 | 0.0280 | No |
| 49 | Ntf3 | 9482 | -0.001 | 0.0101 | No |
| 50 | Mefv | 9522 | -0.002 | 0.0081 | No |
| 51 | Ccdc106 | 9773 | -0.012 | -0.0051 | No |
| 52 | Clstn3 | 9827 | -0.014 | -0.0075 | No |
| 53 | Tcf7l1 | 10102 | -0.026 | -0.0214 | No |
| 54 | Idua | 10301 | -0.034 | -0.0309 | No |
| 55 | Copz2 | 10316 | -0.035 | -0.0303 | No |
| 56 | Sphk2 | 10370 | -0.038 | -0.0317 | No |
| 57 | Gamt | 10596 | -0.049 | -0.0420 | No |
| 58 | Cdkal1 | 10707 | -0.053 | -0.0459 | No |
| 59 | Vps50 | 11278 | -0.077 | -0.0740 | No |
| 60 | Capn9 | 11530 | -0.088 | -0.0843 | No |
| 61 | Epha5 | 11639 | -0.092 | -0.0865 | No |
| 62 | Slc12a3 | 11808 | -0.098 | -0.0919 | No |
| 63 | Slc29a3 | 12085 | -0.111 | -0.1026 | No |
| 64 | Nrip2 | 12197 | -0.116 | -0.1040 | No |
| 65 | Ryr2 | 12702 | -0.140 | -0.1261 | No |
| 66 | Myo15a | 12781 | -0.143 | -0.1247 | No |
| 67 | Sgk1 | 12795 | -0.144 | -0.1197 | No |
| 68 | Tenm2 | 13208 | -0.164 | -0.1357 | No |
| 69 | Ptprj | 13284 | -0.167 | -0.1332 | No |
| 70 | Lfng | 13364 | -0.171 | -0.1308 | No |
| 71 | Skil | 13370 | -0.172 | -0.1243 | No |
| 72 | Coq8a | 13380 | -0.172 | -0.1180 | No |
| 73 | Efhd1 | 13447 | -0.175 | -0.1147 | No |
| 74 | Rgs11 | 13521 | -0.180 | -0.1116 | No |
| 75 | Cpeb3 | 13600 | -0.183 | -0.1087 | No |
| 76 | Bmpr1b | 13745 | -0.191 | -0.1090 | No |
| 77 | Tshb | 13883 | -0.198 | -0.1087 | No |
| 78 | Mx2 | 14281 | -0.221 | -0.1217 | No |
| 79 | Grid2 | 14403 | -0.227 | -0.1193 | No |
| 80 | Mthfr | 14916 | -0.258 | -0.1371 | No |
| 81 | Selenop | 15122 | -0.269 | -0.1377 | No |
| 82 | Tg | 15178 | -0.273 | -0.1300 | No |
| 83 | Gpr19 | 15298 | -0.280 | -0.1254 | No |
| 84 | Tex15 | 15488 | -0.292 | -0.1243 | No |
| 85 | Pde6b | 15540 | -0.296 | -0.1154 | No |
| 86 | Tgm1 | 15668 | -0.305 | -0.1103 | No |
| 87 | Tgfb2 | 16215 | -0.346 | -0.1265 | No |
| 88 | Cyp39a1 | 16497 | -0.369 | -0.1273 | No |
| 89 | Tnni3 | 16584 | -0.379 | -0.1171 | No |
| 90 | Camk1d | 16704 | -0.389 | -0.1082 | No |
| 91 | Fggy | 16885 | -0.409 | -0.1020 | No |
| 92 | Bard1 | 16959 | -0.417 | -0.0895 | No |
| 93 | Gtf3c5 | 17416 | -0.475 | -0.0957 | No |
| 94 | Entpd7 | 17430 | -0.476 | -0.0777 | No |
| 95 | Macroh2a2 | 17924 | -0.573 | -0.0821 | No |
| 96 | Celsr2 | 18075 | -0.633 | -0.0653 | No |
| 97 | Tent5c | 18177 | -0.683 | -0.0439 | No |
| 98 | Rsad2 | 18185 | -0.686 | -0.0173 | No |
| 99 | Synpo | 18282 | -0.747 | 0.0069 | No |

