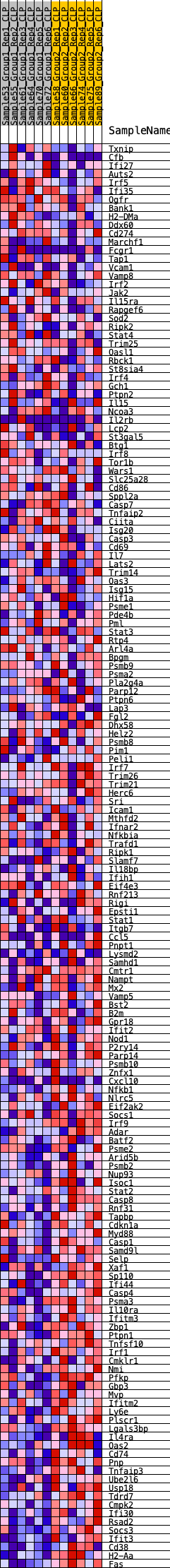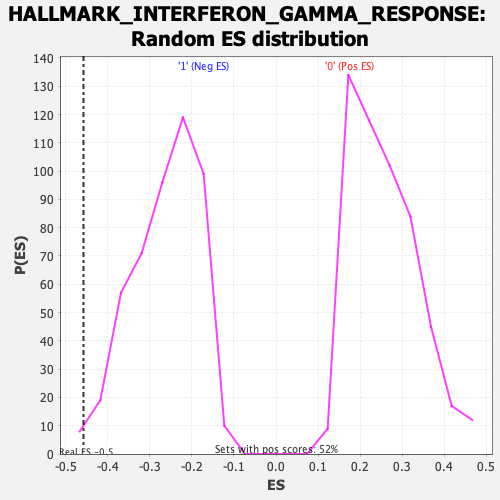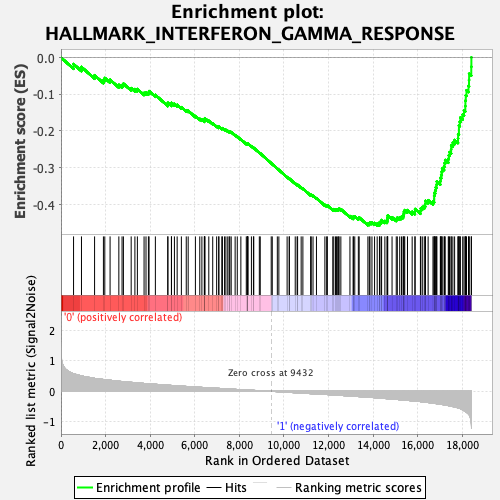
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group1_versus_Group2.Basophil_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_INTERFERON_GAMMA_RESPONSE |
| Enrichment Score (ES) | -0.45743734 |
| Normalized Enrichment Score (NES) | -1.7366178 |
| Nominal p-value | 0.012526096 |
| FDR q-value | 0.04271899 |
| FWER p-Value | 0.061 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Txnip | 563 | 0.576 | -0.0178 | No |
| 2 | Cfb | 918 | 0.508 | -0.0257 | No |
| 3 | Ifi27 | 1506 | 0.424 | -0.0483 | No |
| 4 | Auts2 | 1902 | 0.388 | -0.0611 | No |
| 5 | Irf5 | 1964 | 0.384 | -0.0558 | No |
| 6 | Ifi35 | 2201 | 0.364 | -0.0604 | No |
| 7 | Ogfr | 2594 | 0.331 | -0.0744 | No |
| 8 | Bank1 | 2738 | 0.321 | -0.0750 | No |
| 9 | H2-DMa | 2799 | 0.317 | -0.0711 | No |
| 10 | Ddx60 | 3147 | 0.295 | -0.0834 | No |
| 11 | Cd274 | 3314 | 0.285 | -0.0861 | No |
| 12 | Marchf1 | 3425 | 0.277 | -0.0858 | No |
| 13 | Fcgr1 | 3725 | 0.258 | -0.0964 | No |
| 14 | Tap1 | 3812 | 0.253 | -0.0953 | No |
| 15 | Vcam1 | 3920 | 0.247 | -0.0956 | No |
| 16 | Vamp8 | 3949 | 0.246 | -0.0916 | No |
| 17 | Irf2 | 4232 | 0.231 | -0.1018 | No |
| 18 | Jak2 | 4790 | 0.201 | -0.1278 | No |
| 19 | Il15ra | 4792 | 0.201 | -0.1233 | No |
| 20 | Rapgef6 | 4951 | 0.193 | -0.1275 | No |
| 21 | Sod2 | 4956 | 0.193 | -0.1234 | No |
| 22 | Ripk2 | 5086 | 0.186 | -0.1262 | No |
| 23 | Stat4 | 5209 | 0.180 | -0.1289 | No |
| 24 | Trim25 | 5398 | 0.170 | -0.1353 | No |
| 25 | Oasl1 | 5617 | 0.160 | -0.1436 | No |
| 26 | Rbck1 | 5703 | 0.155 | -0.1448 | No |
| 27 | St8sia4 | 6027 | 0.139 | -0.1593 | No |
| 28 | Irf4 | 6223 | 0.132 | -0.1670 | No |
| 29 | Gch1 | 6318 | 0.128 | -0.1693 | No |
| 30 | Ptpn2 | 6428 | 0.123 | -0.1725 | No |
| 31 | Il15 | 6433 | 0.122 | -0.1699 | No |
| 32 | Ncoa3 | 6445 | 0.122 | -0.1678 | No |
| 33 | Il2rb | 6452 | 0.122 | -0.1653 | No |
| 34 | Lcp2 | 6628 | 0.114 | -0.1723 | No |
| 35 | St3gal5 | 6804 | 0.107 | -0.1795 | No |
| 36 | Btg1 | 6980 | 0.100 | -0.1868 | No |
| 37 | Irf8 | 7072 | 0.096 | -0.1896 | No |
| 38 | Tor1b | 7081 | 0.096 | -0.1879 | No |
| 39 | Wars1 | 7213 | 0.089 | -0.1931 | No |
| 40 | Slc25a28 | 7238 | 0.088 | -0.1924 | No |
| 41 | Cd86 | 7335 | 0.085 | -0.1957 | No |
| 42 | Sppl2a | 7395 | 0.082 | -0.1971 | No |
| 43 | Casp7 | 7479 | 0.078 | -0.1999 | No |
| 44 | Tnfaip2 | 7543 | 0.075 | -0.2016 | No |
| 45 | Ciita | 7571 | 0.074 | -0.2014 | No |
| 46 | Isg20 | 7632 | 0.071 | -0.2031 | No |
| 47 | Casp3 | 7809 | 0.064 | -0.2113 | No |
| 48 | Cd69 | 7905 | 0.060 | -0.2152 | No |
| 49 | Il7 | 8066 | 0.054 | -0.2227 | No |
| 50 | Lats2 | 8309 | 0.044 | -0.2350 | No |
| 51 | Trim14 | 8350 | 0.043 | -0.2362 | No |
| 52 | Oas3 | 8362 | 0.042 | -0.2359 | No |
| 53 | Isg15 | 8372 | 0.042 | -0.2354 | No |
| 54 | Hif1a | 8373 | 0.042 | -0.2345 | No |
| 55 | Psme1 | 8382 | 0.041 | -0.2340 | No |
| 56 | Pde4b | 8539 | 0.036 | -0.2417 | No |
| 57 | Pml | 8630 | 0.032 | -0.2459 | No |
| 58 | Stat3 | 8645 | 0.031 | -0.2460 | No |
| 59 | Rtp4 | 8894 | 0.021 | -0.2591 | No |
| 60 | Arl4a | 8935 | 0.019 | -0.2608 | No |
| 61 | Bpgm | 9427 | 0.000 | -0.2878 | No |
| 62 | Psmb9 | 9474 | -0.001 | -0.2903 | No |
| 63 | Psma2 | 9701 | -0.009 | -0.3024 | No |
| 64 | Pla2g4a | 9770 | -0.012 | -0.3059 | No |
| 65 | Parp12 | 10140 | -0.027 | -0.3255 | No |
| 66 | Ptpn6 | 10235 | -0.031 | -0.3300 | No |
| 67 | Lap3 | 10240 | -0.032 | -0.3295 | No |
| 68 | Fgl2 | 10504 | -0.044 | -0.3429 | No |
| 69 | Dhx58 | 10588 | -0.048 | -0.3463 | No |
| 70 | Helz2 | 10611 | -0.049 | -0.3464 | No |
| 71 | Psmb8 | 10770 | -0.056 | -0.3538 | No |
| 72 | Pim1 | 10847 | -0.060 | -0.3566 | No |
| 73 | Peli1 | 11195 | -0.074 | -0.3740 | No |
| 74 | Irf7 | 11222 | -0.075 | -0.3737 | No |
| 75 | Trim26 | 11302 | -0.078 | -0.3763 | No |
| 76 | Trim21 | 11456 | -0.085 | -0.3827 | No |
| 77 | Herc6 | 11834 | -0.099 | -0.4012 | No |
| 78 | Sri | 11914 | -0.103 | -0.4032 | No |
| 79 | Icam1 | 11945 | -0.104 | -0.4024 | No |
| 80 | Mthfd2 | 12188 | -0.116 | -0.4131 | No |
| 81 | Ifnar2 | 12233 | -0.118 | -0.4128 | No |
| 82 | Nfkbia | 12312 | -0.122 | -0.4143 | No |
| 83 | Trafd1 | 12352 | -0.123 | -0.4137 | No |
| 84 | Ripk1 | 12409 | -0.126 | -0.4139 | No |
| 85 | Slamf7 | 12450 | -0.128 | -0.4132 | No |
| 86 | Il18bp | 12470 | -0.129 | -0.4113 | No |
| 87 | Ifih1 | 12550 | -0.132 | -0.4127 | No |
| 88 | Eif4e3 | 12958 | -0.152 | -0.4315 | No |
| 89 | Rnf213 | 13104 | -0.159 | -0.4359 | No |
| 90 | Rigi | 13111 | -0.159 | -0.4326 | No |
| 91 | Epsti1 | 13174 | -0.162 | -0.4323 | No |
| 92 | Stat1 | 13334 | -0.171 | -0.4372 | No |
| 93 | Itgb7 | 13371 | -0.172 | -0.4352 | No |
| 94 | Ccl5 | 13763 | -0.192 | -0.4523 | No |
| 95 | Pnpt1 | 13826 | -0.195 | -0.4513 | No |
| 96 | Lysmd2 | 13856 | -0.197 | -0.4484 | No |
| 97 | Samhd1 | 13945 | -0.201 | -0.4487 | No |
| 98 | Cmtr1 | 14062 | -0.207 | -0.4504 | No |
| 99 | Nampt | 14179 | -0.214 | -0.4519 | No |
| 100 | Mx2 | 14281 | -0.221 | -0.4524 | Yes |
| 101 | Vamp5 | 14292 | -0.221 | -0.4480 | Yes |
| 102 | Bst2 | 14357 | -0.225 | -0.4464 | Yes |
| 103 | B2m | 14378 | -0.226 | -0.4424 | Yes |
| 104 | Gpr18 | 14524 | -0.235 | -0.4450 | Yes |
| 105 | Ifit2 | 14621 | -0.239 | -0.4448 | Yes |
| 106 | Nod1 | 14636 | -0.240 | -0.4401 | Yes |
| 107 | P2ry14 | 14644 | -0.241 | -0.4351 | Yes |
| 108 | Parp14 | 14652 | -0.241 | -0.4300 | Yes |
| 109 | Psmb10 | 14846 | -0.254 | -0.4348 | Yes |
| 110 | Znfx1 | 15034 | -0.264 | -0.4391 | Yes |
| 111 | Cxcl10 | 15082 | -0.266 | -0.4356 | Yes |
| 112 | Nfkb1 | 15189 | -0.274 | -0.4352 | Yes |
| 113 | Nlrc5 | 15268 | -0.279 | -0.4332 | Yes |
| 114 | Eif2ak2 | 15340 | -0.281 | -0.4307 | Yes |
| 115 | Socs1 | 15369 | -0.283 | -0.4258 | Yes |
| 116 | Irf9 | 15379 | -0.284 | -0.4199 | Yes |
| 117 | Adar | 15415 | -0.287 | -0.4153 | Yes |
| 118 | Batf2 | 15538 | -0.296 | -0.4153 | Yes |
| 119 | Psme2 | 15748 | -0.310 | -0.4197 | Yes |
| 120 | Arid5b | 15872 | -0.320 | -0.4192 | Yes |
| 121 | Psmb2 | 15879 | -0.321 | -0.4123 | Yes |
| 122 | Nup93 | 16118 | -0.336 | -0.4177 | Yes |
| 123 | Isoc1 | 16132 | -0.338 | -0.4107 | Yes |
| 124 | Stat2 | 16202 | -0.344 | -0.4067 | Yes |
| 125 | Casp8 | 16291 | -0.352 | -0.4036 | Yes |
| 126 | Rnf31 | 16338 | -0.356 | -0.3980 | Yes |
| 127 | Tapbp | 16343 | -0.356 | -0.3902 | Yes |
| 128 | Cdkn1a | 16464 | -0.366 | -0.3884 | Yes |
| 129 | Myd88 | 16682 | -0.387 | -0.3915 | Yes |
| 130 | Casp1 | 16734 | -0.392 | -0.3855 | Yes |
| 131 | Samd9l | 16735 | -0.392 | -0.3766 | Yes |
| 132 | Selp | 16745 | -0.393 | -0.3681 | Yes |
| 133 | Xaf1 | 16783 | -0.397 | -0.3612 | Yes |
| 134 | Sp110 | 16806 | -0.400 | -0.3533 | Yes |
| 135 | Ifi44 | 16845 | -0.405 | -0.3462 | Yes |
| 136 | Casp4 | 16854 | -0.406 | -0.3374 | Yes |
| 137 | Psma3 | 17011 | -0.421 | -0.3364 | Yes |
| 138 | Il10ra | 17019 | -0.422 | -0.3273 | Yes |
| 139 | Ifitm3 | 17051 | -0.426 | -0.3193 | Yes |
| 140 | Zbp1 | 17076 | -0.429 | -0.3109 | Yes |
| 141 | Ptpn1 | 17093 | -0.431 | -0.3020 | Yes |
| 142 | Tnfsf10 | 17179 | -0.443 | -0.2966 | Yes |
| 143 | Irf1 | 17199 | -0.445 | -0.2875 | Yes |
| 144 | Cmklr1 | 17229 | -0.448 | -0.2790 | Yes |
| 145 | Nmi | 17368 | -0.468 | -0.2759 | Yes |
| 146 | Pfkp | 17390 | -0.470 | -0.2664 | Yes |
| 147 | Gbp3 | 17424 | -0.476 | -0.2574 | Yes |
| 148 | Mvp | 17495 | -0.488 | -0.2502 | Yes |
| 149 | Ifitm2 | 17499 | -0.489 | -0.2393 | Yes |
| 150 | Ly6e | 17573 | -0.500 | -0.2319 | Yes |
| 151 | Plscr1 | 17652 | -0.512 | -0.2246 | Yes |
| 152 | Lgals3bp | 17804 | -0.541 | -0.2206 | Yes |
| 153 | Il4ra | 17810 | -0.543 | -0.2086 | Yes |
| 154 | Oas2 | 17845 | -0.551 | -0.1980 | Yes |
| 155 | Cd74 | 17846 | -0.551 | -0.1855 | Yes |
| 156 | Pnp | 17885 | -0.560 | -0.1749 | Yes |
| 157 | Tnfaip3 | 17910 | -0.567 | -0.1633 | Yes |
| 158 | Ube2l6 | 18015 | -0.606 | -0.1553 | Yes |
| 159 | Usp18 | 18074 | -0.633 | -0.1441 | Yes |
| 160 | Tdrd7 | 18131 | -0.661 | -0.1322 | Yes |
| 161 | Cmpk2 | 18136 | -0.663 | -0.1174 | Yes |
| 162 | Ifi30 | 18152 | -0.671 | -0.1030 | Yes |
| 163 | Rsad2 | 18185 | -0.686 | -0.0892 | Yes |
| 164 | Socs3 | 18274 | -0.743 | -0.0772 | Yes |
| 165 | Ifit3 | 18298 | -0.759 | -0.0612 | Yes |
| 166 | Cd38 | 18303 | -0.763 | -0.0442 | Yes |
| 167 | H2-Aa | 18399 | -1.068 | -0.0252 | Yes |
| 168 | Fas | 18404 | -1.129 | 0.0002 | Yes |