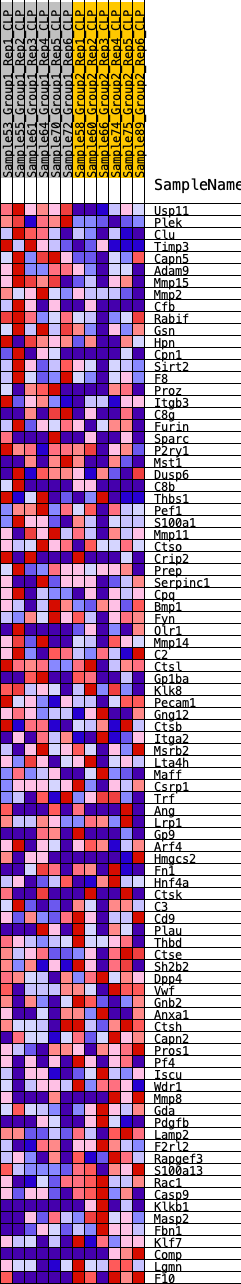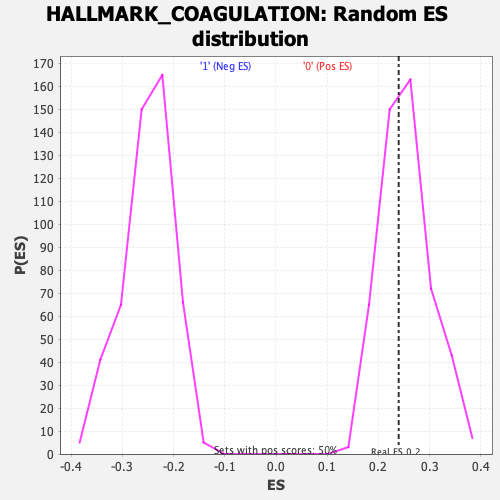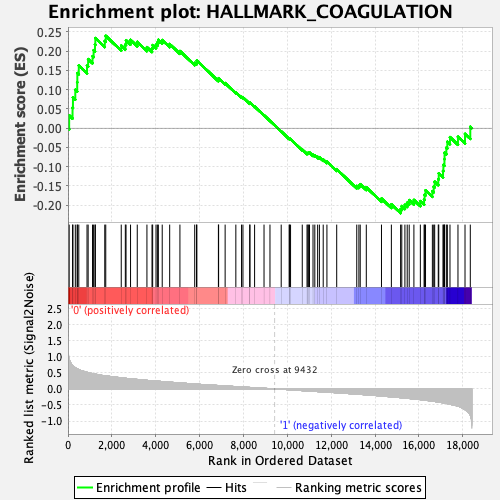
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group1_versus_Group2.Basophil_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_COAGULATION |
| Enrichment Score (ES) | 0.239581 |
| Normalized Enrichment Score (NES) | 0.94653594 |
| Nominal p-value | 0.58449304 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Usp11 | 53 | 0.932 | 0.0333 | Yes |
| 2 | Plek | 207 | 0.727 | 0.0531 | Yes |
| 3 | Clu | 226 | 0.715 | 0.0799 | Yes |
| 4 | Timp3 | 333 | 0.659 | 0.0997 | Yes |
| 5 | Capn5 | 420 | 0.623 | 0.1192 | Yes |
| 6 | Adam9 | 426 | 0.621 | 0.1430 | Yes |
| 7 | Mmp15 | 492 | 0.598 | 0.1627 | Yes |
| 8 | Mmp2 | 867 | 0.515 | 0.1622 | Yes |
| 9 | Cfb | 918 | 0.508 | 0.1792 | Yes |
| 10 | Rabif | 1118 | 0.475 | 0.1868 | Yes |
| 11 | Gsn | 1168 | 0.468 | 0.2023 | Yes |
| 12 | Hpn | 1231 | 0.461 | 0.2168 | Yes |
| 13 | Cpn1 | 1247 | 0.459 | 0.2338 | Yes |
| 14 | Sirt2 | 1676 | 0.408 | 0.2263 | Yes |
| 15 | F8 | 1721 | 0.404 | 0.2396 | Yes |
| 16 | Proz | 2427 | 0.345 | 0.2145 | No |
| 17 | Itgb3 | 2613 | 0.329 | 0.2172 | No |
| 18 | C8g | 2645 | 0.327 | 0.2282 | No |
| 19 | Furin | 2851 | 0.314 | 0.2292 | No |
| 20 | Sparc | 3157 | 0.294 | 0.2239 | No |
| 21 | P2ry1 | 3598 | 0.265 | 0.2102 | No |
| 22 | Mst1 | 3830 | 0.252 | 0.2074 | No |
| 23 | Dusp6 | 3859 | 0.250 | 0.2156 | No |
| 24 | C8b | 4012 | 0.242 | 0.2167 | No |
| 25 | Thbs1 | 4074 | 0.239 | 0.2226 | No |
| 26 | Pef1 | 4119 | 0.238 | 0.2294 | No |
| 27 | S100a1 | 4295 | 0.228 | 0.2287 | No |
| 28 | Mmp11 | 4635 | 0.210 | 0.2184 | No |
| 29 | Ctso | 5102 | 0.185 | 0.2001 | No |
| 30 | Crip2 | 5776 | 0.152 | 0.1693 | No |
| 31 | Prep | 5857 | 0.148 | 0.1707 | No |
| 32 | Serpinc1 | 5875 | 0.147 | 0.1755 | No |
| 33 | Cpq | 6856 | 0.105 | 0.1261 | No |
| 34 | Bmp1 | 6866 | 0.104 | 0.1296 | No |
| 35 | Fyn | 7163 | 0.092 | 0.1170 | No |
| 36 | Olr1 | 7651 | 0.070 | 0.0932 | No |
| 37 | Mmp14 | 7911 | 0.059 | 0.0814 | No |
| 38 | C2 | 7975 | 0.057 | 0.0801 | No |
| 39 | Ctsl | 8286 | 0.045 | 0.0650 | No |
| 40 | Gp1ba | 8293 | 0.045 | 0.0664 | No |
| 41 | Klk8 | 8506 | 0.037 | 0.0562 | No |
| 42 | Pecam1 | 8936 | 0.019 | 0.0336 | No |
| 43 | Gng12 | 9207 | 0.008 | 0.0192 | No |
| 44 | Ctsb | 9719 | -0.010 | -0.0083 | No |
| 45 | Itga2 | 10083 | -0.025 | -0.0272 | No |
| 46 | Msrb2 | 10094 | -0.025 | -0.0268 | No |
| 47 | Lta4h | 10145 | -0.028 | -0.0284 | No |
| 48 | Maff | 10679 | -0.052 | -0.0555 | No |
| 49 | Csrp1 | 10896 | -0.062 | -0.0649 | No |
| 50 | Trf | 10934 | -0.064 | -0.0644 | No |
| 51 | Ang | 10972 | -0.065 | -0.0639 | No |
| 52 | Lrp1 | 11008 | -0.066 | -0.0632 | No |
| 53 | Gp9 | 11169 | -0.072 | -0.0692 | No |
| 54 | Arf4 | 11251 | -0.076 | -0.0706 | No |
| 55 | Hmgcs2 | 11380 | -0.082 | -0.0744 | No |
| 56 | Fn1 | 11460 | -0.085 | -0.0754 | No |
| 57 | Hnf4a | 11634 | -0.092 | -0.0813 | No |
| 58 | Ctsk | 11803 | -0.097 | -0.0867 | No |
| 59 | C3 | 12249 | -0.118 | -0.1064 | No |
| 60 | Cd9 | 13163 | -0.161 | -0.1500 | No |
| 61 | Plau | 13261 | -0.167 | -0.1488 | No |
| 62 | Thbd | 13321 | -0.170 | -0.1454 | No |
| 63 | Ctse | 13598 | -0.183 | -0.1534 | No |
| 64 | Sh2b2 | 14290 | -0.221 | -0.1825 | No |
| 65 | Dpp4 | 14741 | -0.247 | -0.1975 | No |
| 66 | Vwf | 15162 | -0.272 | -0.2099 | No |
| 67 | Gnb2 | 15216 | -0.275 | -0.2021 | No |
| 68 | Anxa1 | 15358 | -0.282 | -0.1988 | No |
| 69 | Ctsh | 15463 | -0.290 | -0.1933 | No |
| 70 | Capn2 | 15554 | -0.297 | -0.1866 | No |
| 71 | Pros1 | 15768 | -0.312 | -0.1862 | No |
| 72 | Pf4 | 16062 | -0.332 | -0.1893 | No |
| 73 | Iscu | 16233 | -0.348 | -0.1851 | No |
| 74 | Wdr1 | 16256 | -0.349 | -0.1727 | No |
| 75 | Mmp8 | 16298 | -0.353 | -0.1612 | No |
| 76 | Gda | 16610 | -0.381 | -0.1634 | No |
| 77 | Pdgfb | 16672 | -0.387 | -0.1517 | No |
| 78 | Lamp2 | 16715 | -0.390 | -0.1389 | No |
| 79 | F2rl2 | 16879 | -0.408 | -0.1319 | No |
| 80 | Rapgef3 | 16901 | -0.411 | -0.1171 | No |
| 81 | S100a13 | 17096 | -0.431 | -0.1110 | No |
| 82 | Rac1 | 17113 | -0.433 | -0.0950 | No |
| 83 | Casp9 | 17161 | -0.441 | -0.0805 | No |
| 84 | Klkb1 | 17166 | -0.442 | -0.0636 | No |
| 85 | Masp2 | 17247 | -0.451 | -0.0504 | No |
| 86 | Fbn1 | 17293 | -0.457 | -0.0351 | No |
| 87 | Klf7 | 17413 | -0.474 | -0.0232 | No |
| 88 | Comp | 17777 | -0.536 | -0.0222 | No |
| 89 | Lgmn | 18098 | -0.643 | -0.0147 | No |
| 90 | F10 | 18339 | -0.815 | 0.0038 | No |