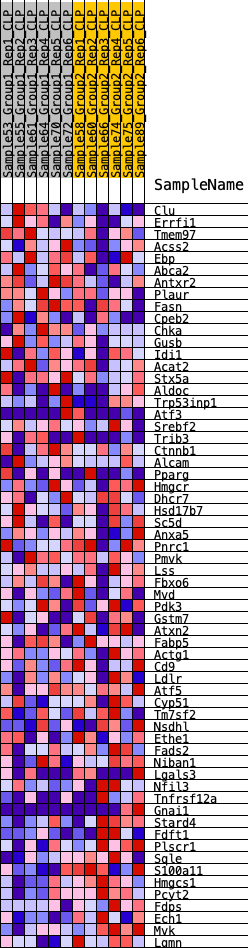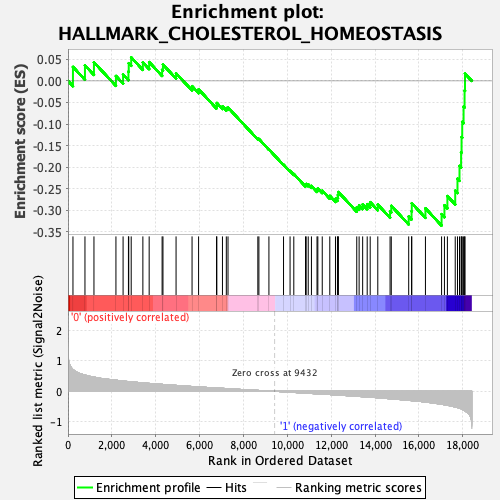
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group1_versus_Group2.Basophil_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_CHOLESTEROL_HOMEOSTASIS |
| Enrichment Score (ES) | -0.33556598 |
| Normalized Enrichment Score (NES) | -1.1985567 |
| Nominal p-value | 0.1820041 |
| FDR q-value | 0.3713167 |
| FWER p-Value | 0.922 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Clu | 226 | 0.715 | 0.0324 | No |
| 2 | Errfi1 | 773 | 0.534 | 0.0361 | No |
| 3 | Tmem97 | 1183 | 0.467 | 0.0431 | No |
| 4 | Acss2 | 2185 | 0.365 | 0.0114 | No |
| 5 | Ebp | 2511 | 0.338 | 0.0149 | No |
| 6 | Abca2 | 2755 | 0.320 | 0.0217 | No |
| 7 | Antxr2 | 2769 | 0.319 | 0.0409 | No |
| 8 | Plaur | 2881 | 0.312 | 0.0545 | No |
| 9 | Fasn | 3413 | 0.278 | 0.0429 | No |
| 10 | Cpeb2 | 3700 | 0.260 | 0.0436 | No |
| 11 | Chka | 4290 | 0.228 | 0.0258 | No |
| 12 | Gusb | 4329 | 0.225 | 0.0378 | No |
| 13 | Idi1 | 4926 | 0.195 | 0.0175 | No |
| 14 | Acat2 | 5655 | 0.157 | -0.0123 | No |
| 15 | Stx5a | 5953 | 0.143 | -0.0195 | No |
| 16 | Aldoc | 6770 | 0.108 | -0.0573 | No |
| 17 | Trp53inp1 | 6785 | 0.108 | -0.0513 | No |
| 18 | Atf3 | 7041 | 0.097 | -0.0591 | No |
| 19 | Srebf2 | 7217 | 0.089 | -0.0631 | No |
| 20 | Trib3 | 7294 | 0.086 | -0.0618 | No |
| 21 | Ctnnb1 | 8654 | 0.031 | -0.1340 | No |
| 22 | Alcam | 8702 | 0.028 | -0.1347 | No |
| 23 | Pparg | 9159 | 0.010 | -0.1590 | No |
| 24 | Hmgcr | 9825 | -0.014 | -0.1943 | No |
| 25 | Dhcr7 | 10123 | -0.027 | -0.2089 | No |
| 26 | Hsd17b7 | 10293 | -0.034 | -0.2159 | No |
| 27 | Sc5d | 10827 | -0.059 | -0.2413 | No |
| 28 | Anxa5 | 10857 | -0.061 | -0.2391 | No |
| 29 | Pnrc1 | 10951 | -0.064 | -0.2401 | No |
| 30 | Pmvk | 11097 | -0.070 | -0.2436 | No |
| 31 | Lss | 11352 | -0.081 | -0.2524 | No |
| 32 | Fbxo6 | 11389 | -0.082 | -0.2492 | No |
| 33 | Mvd | 11590 | -0.090 | -0.2545 | No |
| 34 | Pdk3 | 11934 | -0.104 | -0.2667 | No |
| 35 | Gstm7 | 12193 | -0.116 | -0.2735 | No |
| 36 | Atxn2 | 12283 | -0.120 | -0.2708 | No |
| 37 | Fabp5 | 12306 | -0.121 | -0.2644 | No |
| 38 | Actg1 | 12325 | -0.122 | -0.2577 | No |
| 39 | Cd9 | 13163 | -0.161 | -0.2933 | No |
| 40 | Ldlr | 13271 | -0.167 | -0.2886 | No |
| 41 | Atf5 | 13432 | -0.175 | -0.2864 | No |
| 42 | Cyp51 | 13639 | -0.186 | -0.2860 | No |
| 43 | Tm7sf2 | 13776 | -0.193 | -0.2813 | No |
| 44 | Nsdhl | 14122 | -0.211 | -0.2869 | No |
| 45 | Ethe1 | 14682 | -0.243 | -0.3022 | No |
| 46 | Fads2 | 14737 | -0.247 | -0.2897 | No |
| 47 | Niban1 | 15531 | -0.295 | -0.3144 | Yes |
| 48 | Lgals3 | 15661 | -0.305 | -0.3024 | Yes |
| 49 | Nfil3 | 15672 | -0.306 | -0.2838 | Yes |
| 50 | Tnfrsf12a | 16294 | -0.353 | -0.2955 | Yes |
| 51 | Gnai1 | 17030 | -0.423 | -0.3090 | Yes |
| 52 | Stard4 | 17160 | -0.441 | -0.2885 | Yes |
| 53 | Fdft1 | 17295 | -0.457 | -0.2672 | Yes |
| 54 | Plscr1 | 17652 | -0.512 | -0.2545 | Yes |
| 55 | Sqle | 17755 | -0.531 | -0.2268 | Yes |
| 56 | S100a11 | 17847 | -0.551 | -0.1972 | Yes |
| 57 | Hmgcs1 | 17920 | -0.570 | -0.1654 | Yes |
| 58 | Pcyt2 | 17941 | -0.578 | -0.1303 | Yes |
| 59 | Fdps | 17971 | -0.591 | -0.0949 | Yes |
| 60 | Ech1 | 18033 | -0.615 | -0.0597 | Yes |
| 61 | Mvk | 18078 | -0.635 | -0.0223 | Yes |
| 62 | Lgmn | 18098 | -0.643 | 0.0169 | Yes |