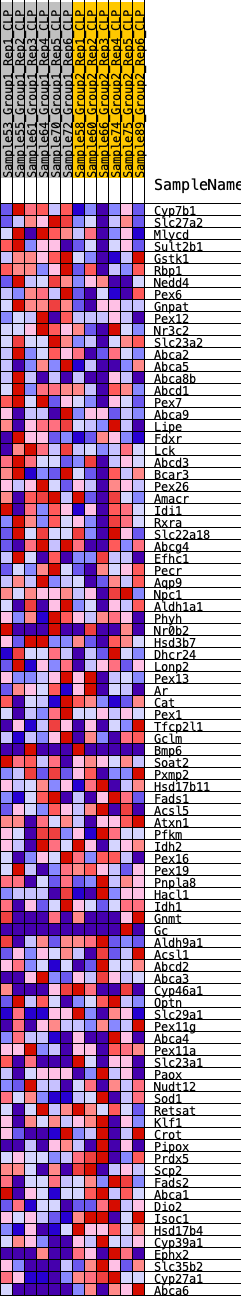
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group1_versus_Group2.Basophil_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 0 |
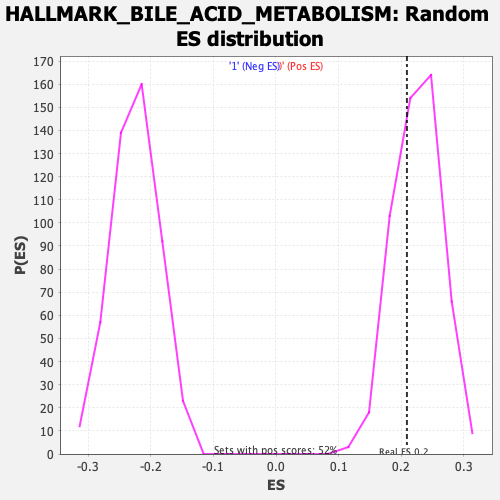
| GeneSet | HALLMARK_BILE_ACID_METABOLISM |
| Enrichment Score (ES) | 0.20948632 |
| Normalized Enrichment Score (NES) | 0.9249091 |
| Nominal p-value | 0.65377176 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Cyp7b1 | 100 | 0.835 | 0.0366 | Yes |
| 2 | Slc27a2 | 613 | 0.566 | 0.0373 | Yes |
| 3 | Mlycd | 935 | 0.504 | 0.0451 | Yes |
| 4 | Sult2b1 | 1121 | 0.475 | 0.0590 | Yes |
| 5 | Gstk1 | 1213 | 0.463 | 0.0774 | Yes |
| 6 | Rbp1 | 1237 | 0.461 | 0.0993 | Yes |
| 7 | Nedd4 | 1476 | 0.429 | 0.1080 | Yes |
| 8 | Pex6 | 1539 | 0.421 | 0.1258 | Yes |
| 9 | Gnpat | 1564 | 0.418 | 0.1456 | Yes |
| 10 | Pex12 | 2231 | 0.362 | 0.1274 | Yes |
| 11 | Nr3c2 | 2499 | 0.340 | 0.1300 | Yes |
| 12 | Slc23a2 | 2549 | 0.335 | 0.1442 | Yes |
| 13 | Abca2 | 2755 | 0.320 | 0.1492 | Yes |
| 14 | Abca5 | 2860 | 0.314 | 0.1593 | Yes |
| 15 | Abca8b | 2912 | 0.310 | 0.1722 | Yes |
| 16 | Abcd1 | 3079 | 0.299 | 0.1782 | Yes |
| 17 | Pex7 | 3655 | 0.261 | 0.1599 | Yes |
| 18 | Abca9 | 3806 | 0.253 | 0.1645 | Yes |
| 19 | Lipe | 3815 | 0.253 | 0.1768 | Yes |
| 20 | Fdxr | 3824 | 0.253 | 0.1891 | Yes |
| 21 | Lck | 3863 | 0.250 | 0.1997 | Yes |
| 22 | Abcd3 | 4025 | 0.241 | 0.2030 | Yes |
| 23 | Bcar3 | 4127 | 0.237 | 0.2095 | Yes |
| 24 | Pex26 | 4431 | 0.220 | 0.2041 | No |
| 25 | Amacr | 4570 | 0.214 | 0.2073 | No |
| 26 | Idi1 | 4926 | 0.195 | 0.1977 | No |
| 27 | Rxra | 5062 | 0.187 | 0.1998 | No |
| 28 | Slc22a18 | 5624 | 0.159 | 0.1772 | No |
| 29 | Abcg4 | 5628 | 0.159 | 0.1851 | No |
| 30 | Efhc1 | 5932 | 0.144 | 0.1758 | No |
| 31 | Pecr | 5943 | 0.143 | 0.1825 | No |
| 32 | Aqp9 | 6088 | 0.137 | 0.1815 | No |
| 33 | Npc1 | 6180 | 0.134 | 0.1833 | No |
| 34 | Aldh1a1 | 6188 | 0.134 | 0.1896 | No |
| 35 | Phyh | 6196 | 0.133 | 0.1960 | No |
| 36 | Nr0b2 | 6235 | 0.132 | 0.2005 | No |
| 37 | Hsd3b7 | 6309 | 0.128 | 0.2030 | No |
| 38 | Dhcr24 | 6850 | 0.105 | 0.1788 | No |
| 39 | Lonp2 | 7246 | 0.088 | 0.1617 | No |
| 40 | Pex13 | 7578 | 0.074 | 0.1473 | No |
| 41 | Ar | 7669 | 0.070 | 0.1459 | No |
| 42 | Cat | 8683 | 0.029 | 0.0921 | No |
| 43 | Pex1 | 8767 | 0.026 | 0.0889 | No |
| 44 | Tfcp2l1 | 8798 | 0.024 | 0.0885 | No |
| 45 | Gclm | 8966 | 0.018 | 0.0803 | No |
| 46 | Bmp6 | 9220 | 0.008 | 0.0668 | No |
| 47 | Soat2 | 9629 | -0.006 | 0.0449 | No |
| 48 | Pxmp2 | 9690 | -0.008 | 0.0420 | No |
| 49 | Hsd17b11 | 9799 | -0.013 | 0.0368 | No |
| 50 | Fads1 | 10009 | -0.021 | 0.0265 | No |
| 51 | Acsl5 | 10049 | -0.024 | 0.0255 | No |
| 52 | Atxn1 | 10153 | -0.028 | 0.0213 | No |
| 53 | Pfkm | 10283 | -0.034 | 0.0160 | No |
| 54 | Idh2 | 10590 | -0.048 | 0.0017 | No |
| 55 | Pex16 | 10674 | -0.052 | -0.0002 | No |
| 56 | Pex19 | 10736 | -0.055 | -0.0008 | No |
| 57 | Pnpla8 | 11047 | -0.068 | -0.0143 | No |
| 58 | Hacl1 | 11051 | -0.068 | -0.0110 | No |
| 59 | Idh1 | 11326 | -0.080 | -0.0219 | No |
| 60 | Gnmt | 11616 | -0.091 | -0.0331 | No |
| 61 | Gc | 11717 | -0.095 | -0.0338 | No |
| 62 | Aldh9a1 | 12096 | -0.112 | -0.0488 | No |
| 63 | Acsl1 | 12141 | -0.114 | -0.0454 | No |
| 64 | Abcd2 | 12445 | -0.127 | -0.0556 | No |
| 65 | Abca3 | 12461 | -0.128 | -0.0499 | No |
| 66 | Cyp46a1 | 12471 | -0.129 | -0.0439 | No |
| 67 | Optn | 12477 | -0.129 | -0.0377 | No |
| 68 | Slc29a1 | 12773 | -0.143 | -0.0466 | No |
| 69 | Pex11g | 12787 | -0.144 | -0.0400 | No |
| 70 | Abca4 | 12887 | -0.148 | -0.0380 | No |
| 71 | Pex11a | 12916 | -0.150 | -0.0319 | No |
| 72 | Slc23a1 | 12941 | -0.151 | -0.0256 | No |
| 73 | Paox | 12988 | -0.154 | -0.0204 | No |
| 74 | Nudt12 | 13210 | -0.164 | -0.0242 | No |
| 75 | Sod1 | 13368 | -0.171 | -0.0241 | No |
| 76 | Retsat | 13642 | -0.186 | -0.0296 | No |
| 77 | Klf1 | 13699 | -0.189 | -0.0231 | No |
| 78 | Crot | 13780 | -0.193 | -0.0178 | No |
| 79 | Pipox | 13878 | -0.198 | -0.0131 | No |
| 80 | Prdx5 | 14464 | -0.231 | -0.0334 | No |
| 81 | Scp2 | 14620 | -0.239 | -0.0298 | No |
| 82 | Fads2 | 14737 | -0.247 | -0.0237 | No |
| 83 | Abca1 | 15780 | -0.313 | -0.0648 | No |
| 84 | Dio2 | 16010 | -0.329 | -0.0607 | No |
| 85 | Isoc1 | 16132 | -0.338 | -0.0502 | No |
| 86 | Hsd17b4 | 16303 | -0.353 | -0.0417 | No |
| 87 | Cyp39a1 | 16497 | -0.369 | -0.0336 | No |
| 88 | Ephx2 | 17944 | -0.579 | -0.0834 | No |
| 89 | Slc35b2 | 18119 | -0.654 | -0.0599 | No |
| 90 | Cyp27a1 | 18249 | -0.722 | -0.0305 | No |
| 91 | Abca6 | 18310 | -0.776 | 0.0054 | No |