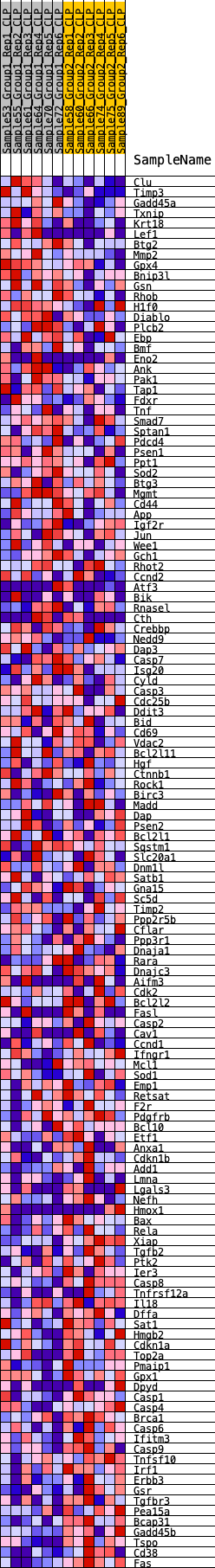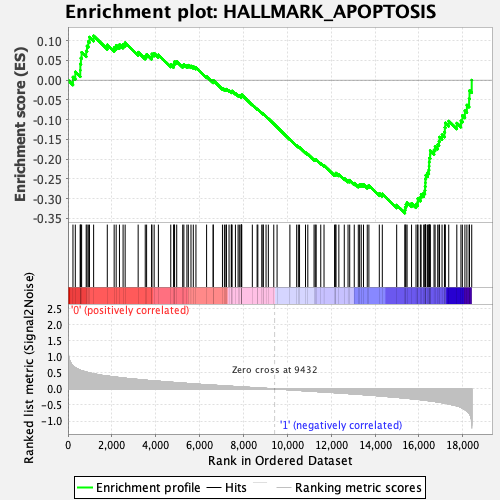
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group1_versus_Group2.Basophil_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_APOPTOSIS |
| Enrichment Score (ES) | -0.3366801 |
| Normalized Enrichment Score (NES) | -1.4999406 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.14063235 |
| FWER p-Value | 0.375 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Clu | 226 | 0.715 | 0.0079 | No |
| 2 | Timp3 | 333 | 0.659 | 0.0209 | No |
| 3 | Gadd45a | 559 | 0.578 | 0.0250 | No |
| 4 | Txnip | 563 | 0.576 | 0.0412 | No |
| 5 | Krt18 | 589 | 0.571 | 0.0560 | No |
| 6 | Lef1 | 621 | 0.565 | 0.0704 | No |
| 7 | Btg2 | 829 | 0.524 | 0.0740 | No |
| 8 | Mmp2 | 867 | 0.515 | 0.0866 | No |
| 9 | Gpx4 | 925 | 0.507 | 0.0979 | No |
| 10 | Bnip3l | 979 | 0.497 | 0.1091 | No |
| 11 | Gsn | 1168 | 0.468 | 0.1121 | No |
| 12 | Rhob | 1791 | 0.398 | 0.0894 | No |
| 13 | H1f0 | 2104 | 0.371 | 0.0828 | No |
| 14 | Diablo | 2196 | 0.364 | 0.0882 | No |
| 15 | Plcb2 | 2346 | 0.351 | 0.0900 | No |
| 16 | Ebp | 2511 | 0.338 | 0.0907 | No |
| 17 | Bmf | 2605 | 0.330 | 0.0950 | No |
| 18 | Eno2 | 3198 | 0.291 | 0.0708 | No |
| 19 | Ank | 3521 | 0.271 | 0.0609 | No |
| 20 | Pak1 | 3583 | 0.267 | 0.0652 | No |
| 21 | Tap1 | 3812 | 0.253 | 0.0599 | No |
| 22 | Fdxr | 3824 | 0.253 | 0.0665 | No |
| 23 | Tnf | 3927 | 0.246 | 0.0679 | No |
| 24 | Smad7 | 4120 | 0.238 | 0.0641 | No |
| 25 | Sptan1 | 4684 | 0.208 | 0.0392 | No |
| 26 | Pdcd4 | 4815 | 0.200 | 0.0378 | No |
| 27 | Psen1 | 4836 | 0.199 | 0.0423 | No |
| 28 | Ppt1 | 4858 | 0.198 | 0.0468 | No |
| 29 | Sod2 | 4956 | 0.193 | 0.0470 | No |
| 30 | Btg3 | 5229 | 0.179 | 0.0372 | No |
| 31 | Mgmt | 5282 | 0.176 | 0.0394 | No |
| 32 | Cd44 | 5421 | 0.169 | 0.0366 | No |
| 33 | App | 5485 | 0.166 | 0.0379 | No |
| 34 | Igf2r | 5606 | 0.160 | 0.0359 | No |
| 35 | Jun | 5712 | 0.155 | 0.0345 | No |
| 36 | Wee1 | 5832 | 0.149 | 0.0323 | No |
| 37 | Gch1 | 6318 | 0.128 | 0.0094 | No |
| 38 | Rhot2 | 6618 | 0.115 | -0.0037 | No |
| 39 | Ccnd2 | 6620 | 0.115 | -0.0005 | No |
| 40 | Atf3 | 7041 | 0.097 | -0.0207 | No |
| 41 | Bik | 7130 | 0.094 | -0.0229 | No |
| 42 | Rnasel | 7199 | 0.090 | -0.0240 | No |
| 43 | Cth | 7223 | 0.089 | -0.0228 | No |
| 44 | Crebbp | 7341 | 0.084 | -0.0268 | No |
| 45 | Nedd9 | 7442 | 0.080 | -0.0300 | No |
| 46 | Dap3 | 7455 | 0.079 | -0.0284 | No |
| 47 | Casp7 | 7479 | 0.078 | -0.0274 | No |
| 48 | Isg20 | 7632 | 0.071 | -0.0337 | No |
| 49 | Cyld | 7753 | 0.066 | -0.0384 | No |
| 50 | Casp3 | 7809 | 0.064 | -0.0396 | No |
| 51 | Cdc25b | 7897 | 0.060 | -0.0426 | No |
| 52 | Ddit3 | 7898 | 0.060 | -0.0409 | No |
| 53 | Bid | 7902 | 0.060 | -0.0394 | No |
| 54 | Cd69 | 7905 | 0.060 | -0.0378 | No |
| 55 | Vdac2 | 7921 | 0.059 | -0.0369 | No |
| 56 | Bcl2l11 | 8403 | 0.041 | -0.0621 | No |
| 57 | Hgf | 8621 | 0.032 | -0.0731 | No |
| 58 | Ctnnb1 | 8654 | 0.031 | -0.0739 | No |
| 59 | Rock1 | 8833 | 0.023 | -0.0830 | No |
| 60 | Birc3 | 8890 | 0.021 | -0.0855 | No |
| 61 | Madd | 8901 | 0.020 | -0.0855 | No |
| 62 | Dap | 9028 | 0.016 | -0.0919 | No |
| 63 | Psen2 | 9133 | 0.012 | -0.0973 | No |
| 64 | Bcl2l1 | 9380 | 0.002 | -0.1107 | No |
| 65 | Sqstm1 | 9531 | -0.003 | -0.1188 | No |
| 66 | Slc20a1 | 10115 | -0.026 | -0.1500 | No |
| 67 | Dnm1l | 10424 | -0.041 | -0.1657 | No |
| 68 | Satb1 | 10500 | -0.044 | -0.1685 | No |
| 69 | Gna15 | 10555 | -0.047 | -0.1701 | No |
| 70 | Sc5d | 10827 | -0.059 | -0.1833 | No |
| 71 | Timp2 | 10932 | -0.064 | -0.1872 | No |
| 72 | Ppp2r5b | 11217 | -0.075 | -0.2006 | No |
| 73 | Cflar | 11272 | -0.077 | -0.2014 | No |
| 74 | Ppp3r1 | 11313 | -0.079 | -0.2013 | No |
| 75 | Dnaja1 | 11522 | -0.088 | -0.2102 | No |
| 76 | Rara | 11671 | -0.094 | -0.2156 | No |
| 77 | Dnajc3 | 12151 | -0.114 | -0.2386 | No |
| 78 | Aifm3 | 12205 | -0.116 | -0.2382 | No |
| 79 | Cdk2 | 12212 | -0.117 | -0.2352 | No |
| 80 | Bcl2l2 | 12347 | -0.123 | -0.2390 | No |
| 81 | Fasl | 12594 | -0.134 | -0.2487 | No |
| 82 | Casp2 | 12766 | -0.142 | -0.2540 | No |
| 83 | Cav1 | 12842 | -0.146 | -0.2539 | No |
| 84 | Ccnd1 | 13049 | -0.156 | -0.2608 | No |
| 85 | Ifngr1 | 13233 | -0.165 | -0.2661 | No |
| 86 | Mcl1 | 13294 | -0.168 | -0.2646 | No |
| 87 | Sod1 | 13368 | -0.171 | -0.2637 | No |
| 88 | Emp1 | 13470 | -0.176 | -0.2642 | No |
| 89 | Retsat | 13642 | -0.186 | -0.2683 | No |
| 90 | F2r | 13716 | -0.190 | -0.2669 | No |
| 91 | Pdgfrb | 14191 | -0.214 | -0.2868 | No |
| 92 | Bcl10 | 14328 | -0.223 | -0.2879 | No |
| 93 | Etf1 | 14983 | -0.262 | -0.3162 | No |
| 94 | Anxa1 | 15358 | -0.282 | -0.3287 | Yes |
| 95 | Cdkn1b | 15371 | -0.283 | -0.3213 | Yes |
| 96 | Add1 | 15406 | -0.286 | -0.3150 | Yes |
| 97 | Lmna | 15457 | -0.289 | -0.3095 | Yes |
| 98 | Lgals3 | 15661 | -0.305 | -0.3120 | Yes |
| 99 | Nefh | 15865 | -0.319 | -0.3140 | Yes |
| 100 | Hmox1 | 15944 | -0.325 | -0.3090 | Yes |
| 101 | Bax | 15948 | -0.325 | -0.3000 | Yes |
| 102 | Rela | 16063 | -0.332 | -0.2968 | Yes |
| 103 | Xiap | 16096 | -0.335 | -0.2890 | Yes |
| 104 | Tgfb2 | 16215 | -0.346 | -0.2856 | Yes |
| 105 | Ptk2 | 16270 | -0.350 | -0.2786 | Yes |
| 106 | Ier3 | 16279 | -0.351 | -0.2691 | Yes |
| 107 | Casp8 | 16291 | -0.352 | -0.2597 | Yes |
| 108 | Tnfrsf12a | 16294 | -0.353 | -0.2498 | Yes |
| 109 | Il18 | 16312 | -0.354 | -0.2406 | Yes |
| 110 | Dffa | 16391 | -0.360 | -0.2346 | Yes |
| 111 | Sat1 | 16449 | -0.365 | -0.2274 | Yes |
| 112 | Hmgb2 | 16459 | -0.366 | -0.2175 | Yes |
| 113 | Cdkn1a | 16464 | -0.366 | -0.2073 | Yes |
| 114 | Top2a | 16471 | -0.367 | -0.1972 | Yes |
| 115 | Pmaip1 | 16509 | -0.370 | -0.1887 | Yes |
| 116 | Gpx1 | 16511 | -0.370 | -0.1782 | Yes |
| 117 | Dpyd | 16686 | -0.388 | -0.1767 | Yes |
| 118 | Casp1 | 16734 | -0.392 | -0.1681 | Yes |
| 119 | Casp4 | 16854 | -0.406 | -0.1631 | Yes |
| 120 | Brca1 | 16912 | -0.412 | -0.1545 | Yes |
| 121 | Casp6 | 16937 | -0.415 | -0.1441 | Yes |
| 122 | Ifitm3 | 17051 | -0.426 | -0.1381 | Yes |
| 123 | Casp9 | 17161 | -0.441 | -0.1316 | Yes |
| 124 | Tnfsf10 | 17179 | -0.443 | -0.1199 | Yes |
| 125 | Irf1 | 17199 | -0.445 | -0.1083 | Yes |
| 126 | Erbb3 | 17356 | -0.466 | -0.1036 | Yes |
| 127 | Gsr | 17724 | -0.526 | -0.1087 | Yes |
| 128 | Tgfbr3 | 17913 | -0.568 | -0.1028 | Yes |
| 129 | Pea15a | 17977 | -0.594 | -0.0894 | Yes |
| 130 | Bcap31 | 18093 | -0.641 | -0.0775 | Yes |
| 131 | Gadd45b | 18184 | -0.686 | -0.0629 | Yes |
| 132 | Tspo | 18284 | -0.748 | -0.0471 | Yes |
| 133 | Cd38 | 18303 | -0.763 | -0.0264 | Yes |
| 134 | Fas | 18404 | -1.129 | 0.0002 | Yes |