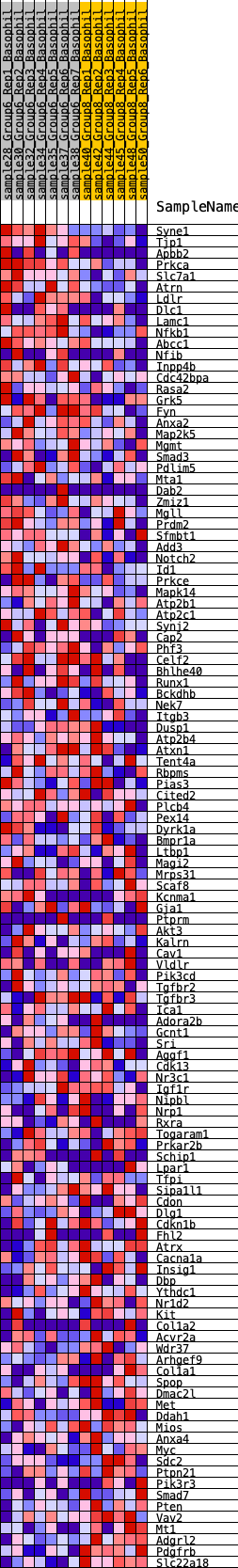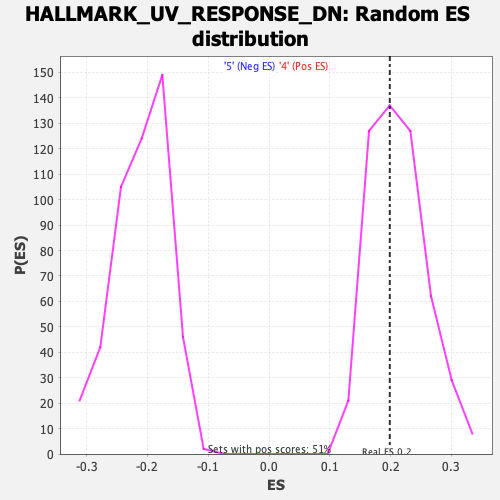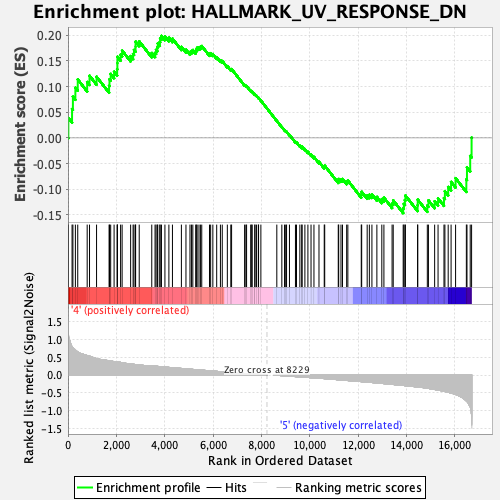
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group6_versus_Group8.Basophil_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | Basophil_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 4 |
| GeneSet | HALLMARK_UV_RESPONSE_DN |
| Enrichment Score (ES) | 0.19858775 |
| Normalized Enrichment Score (NES) | 0.9375293 |
| Nominal p-value | 0.5812133 |
| FDR q-value | 0.8074395 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Syne1 | 15 | 1.176 | 0.0388 | Yes |
| 2 | Tjp1 | 168 | 0.794 | 0.0564 | Yes |
| 3 | Apbb2 | 203 | 0.768 | 0.0802 | Yes |
| 4 | Prkca | 305 | 0.700 | 0.0978 | Yes |
| 5 | Slc7a1 | 403 | 0.647 | 0.1137 | Yes |
| 6 | Atrn | 793 | 0.550 | 0.1088 | Yes |
| 7 | Ldlr | 890 | 0.531 | 0.1210 | Yes |
| 8 | Dlc1 | 1181 | 0.461 | 0.1190 | Yes |
| 9 | Lamc1 | 1699 | 0.405 | 0.1015 | Yes |
| 10 | Nfkb1 | 1716 | 0.403 | 0.1142 | Yes |
| 11 | Abcc1 | 1765 | 0.394 | 0.1246 | Yes |
| 12 | Nfib | 1906 | 0.379 | 0.1289 | Yes |
| 13 | Inpp4b | 2032 | 0.368 | 0.1338 | Yes |
| 14 | Cdc42bpa | 2042 | 0.367 | 0.1456 | Yes |
| 15 | Rasa2 | 2045 | 0.367 | 0.1579 | Yes |
| 16 | Grk5 | 2175 | 0.353 | 0.1621 | Yes |
| 17 | Fyn | 2238 | 0.346 | 0.1700 | Yes |
| 18 | Anxa2 | 2590 | 0.310 | 0.1593 | Yes |
| 19 | Map2k5 | 2692 | 0.303 | 0.1634 | Yes |
| 20 | Mgmt | 2730 | 0.300 | 0.1713 | Yes |
| 21 | Smad3 | 2793 | 0.295 | 0.1775 | Yes |
| 22 | Pdlim5 | 2795 | 0.294 | 0.1874 | Yes |
| 23 | Mta1 | 2945 | 0.282 | 0.1879 | Yes |
| 24 | Dab2 | 3464 | 0.255 | 0.1653 | Yes |
| 25 | Zmiz1 | 3600 | 0.251 | 0.1656 | Yes |
| 26 | Mgll | 3642 | 0.248 | 0.1715 | Yes |
| 27 | Prdm2 | 3695 | 0.244 | 0.1766 | Yes |
| 28 | Sfmbt1 | 3712 | 0.243 | 0.1839 | Yes |
| 29 | Add3 | 3797 | 0.237 | 0.1868 | Yes |
| 30 | Notch2 | 3810 | 0.236 | 0.1940 | Yes |
| 31 | Id1 | 3865 | 0.232 | 0.1986 | Yes |
| 32 | Prkce | 4008 | 0.226 | 0.1977 | No |
| 33 | Mapk14 | 4172 | 0.215 | 0.1951 | No |
| 34 | Atp2b1 | 4319 | 0.205 | 0.1932 | No |
| 35 | Atp2c1 | 4689 | 0.185 | 0.1772 | No |
| 36 | Synj2 | 4878 | 0.174 | 0.1717 | No |
| 37 | Cap2 | 5044 | 0.164 | 0.1673 | No |
| 38 | Phf3 | 5105 | 0.161 | 0.1691 | No |
| 39 | Celf2 | 5162 | 0.158 | 0.1711 | No |
| 40 | Bhlhe40 | 5281 | 0.152 | 0.1691 | No |
| 41 | Runx1 | 5308 | 0.151 | 0.1726 | No |
| 42 | Bckdhb | 5340 | 0.149 | 0.1758 | No |
| 43 | Nek7 | 5416 | 0.145 | 0.1762 | No |
| 44 | Itgb3 | 5476 | 0.142 | 0.1774 | No |
| 45 | Dusp1 | 5531 | 0.139 | 0.1789 | No |
| 46 | Atp2b4 | 5851 | 0.121 | 0.1637 | No |
| 47 | Atxn1 | 5899 | 0.118 | 0.1648 | No |
| 48 | Tent4a | 5990 | 0.112 | 0.1632 | No |
| 49 | Rbpms | 6149 | 0.104 | 0.1572 | No |
| 50 | Pias3 | 6303 | 0.095 | 0.1511 | No |
| 51 | Cited2 | 6379 | 0.091 | 0.1497 | No |
| 52 | Plcb4 | 6589 | 0.079 | 0.1397 | No |
| 53 | Pex14 | 6733 | 0.071 | 0.1335 | No |
| 54 | Dyrk1a | 6765 | 0.070 | 0.1340 | No |
| 55 | Bmpr1a | 7303 | 0.043 | 0.1031 | No |
| 56 | Ltbp1 | 7355 | 0.040 | 0.1014 | No |
| 57 | Magi2 | 7385 | 0.039 | 0.1009 | No |
| 58 | Mrps31 | 7555 | 0.032 | 0.0918 | No |
| 59 | Scaf8 | 7597 | 0.030 | 0.0903 | No |
| 60 | Kcnma1 | 7615 | 0.029 | 0.0903 | No |
| 61 | Gja1 | 7726 | 0.024 | 0.0844 | No |
| 62 | Ptprm | 7730 | 0.024 | 0.0851 | No |
| 63 | Akt3 | 7793 | 0.021 | 0.0820 | No |
| 64 | Kalrn | 7870 | 0.016 | 0.0780 | No |
| 65 | Cav1 | 7880 | 0.016 | 0.0780 | No |
| 66 | Vldlr | 7976 | 0.011 | 0.0726 | No |
| 67 | Pik3cd | 8635 | -0.006 | 0.0332 | No |
| 68 | Tgfbr2 | 8843 | -0.016 | 0.0212 | No |
| 69 | Tgfbr3 | 8955 | -0.022 | 0.0152 | No |
| 70 | Ica1 | 9004 | -0.024 | 0.0131 | No |
| 71 | Adora2b | 9038 | -0.026 | 0.0120 | No |
| 72 | Gcnt1 | 9158 | -0.031 | 0.0059 | No |
| 73 | Sri | 9406 | -0.042 | -0.0076 | No |
| 74 | Aggf1 | 9449 | -0.044 | -0.0086 | No |
| 75 | Cdk13 | 9588 | -0.050 | -0.0153 | No |
| 76 | Nr3c1 | 9666 | -0.054 | -0.0181 | No |
| 77 | Igf1r | 9680 | -0.055 | -0.0170 | No |
| 78 | Nipbl | 9796 | -0.060 | -0.0219 | No |
| 79 | Nrp1 | 9921 | -0.066 | -0.0271 | No |
| 80 | Rxra | 10050 | -0.073 | -0.0324 | No |
| 81 | Togaram1 | 10171 | -0.078 | -0.0370 | No |
| 82 | Prkar2b | 10379 | -0.087 | -0.0466 | No |
| 83 | Schip1 | 10596 | -0.098 | -0.0563 | No |
| 84 | Lpar1 | 10616 | -0.099 | -0.0541 | No |
| 85 | Tfpi | 11176 | -0.131 | -0.0833 | No |
| 86 | Sipa1l1 | 11194 | -0.132 | -0.0799 | No |
| 87 | Cdon | 11289 | -0.138 | -0.0809 | No |
| 88 | Dlg1 | 11351 | -0.140 | -0.0799 | No |
| 89 | Cdkn1b | 11524 | -0.150 | -0.0852 | No |
| 90 | Fhl2 | 11573 | -0.153 | -0.0829 | No |
| 91 | Atrx | 12127 | -0.183 | -0.1101 | No |
| 92 | Cacna1a | 12145 | -0.184 | -0.1049 | No |
| 93 | Insig1 | 12372 | -0.195 | -0.1119 | No |
| 94 | Dbp | 12462 | -0.200 | -0.1105 | No |
| 95 | Ythdc1 | 12568 | -0.208 | -0.1099 | No |
| 96 | Nr1d2 | 12773 | -0.220 | -0.1147 | No |
| 97 | Kit | 12977 | -0.234 | -0.1191 | No |
| 98 | Col1a2 | 13064 | -0.239 | -0.1162 | No |
| 99 | Acvr2a | 13402 | -0.261 | -0.1277 | No |
| 100 | Wdr37 | 13449 | -0.264 | -0.1216 | No |
| 101 | Arhgef9 | 13861 | -0.290 | -0.1366 | No |
| 102 | Col1a1 | 13888 | -0.292 | -0.1283 | No |
| 103 | Spop | 13932 | -0.296 | -0.1209 | No |
| 104 | Dmac2l | 13953 | -0.298 | -0.1121 | No |
| 105 | Met | 14457 | -0.335 | -0.1311 | No |
| 106 | Ddah1 | 14463 | -0.335 | -0.1201 | No |
| 107 | Mios | 14859 | -0.370 | -0.1314 | No |
| 108 | Anxa4 | 14905 | -0.375 | -0.1215 | No |
| 109 | Myc | 15162 | -0.404 | -0.1233 | No |
| 110 | Sdc2 | 15303 | -0.419 | -0.1176 | No |
| 111 | Ptpn21 | 15548 | -0.454 | -0.1170 | No |
| 112 | Pik3r3 | 15586 | -0.461 | -0.1037 | No |
| 113 | Smad7 | 15722 | -0.485 | -0.0954 | No |
| 114 | Pten | 15842 | -0.504 | -0.0856 | No |
| 115 | Vav2 | 16028 | -0.543 | -0.0784 | No |
| 116 | Mt1 | 16473 | -0.727 | -0.0807 | No |
| 117 | Adgrl2 | 16499 | -0.743 | -0.0571 | No |
| 118 | Pdgfrb | 16633 | -0.894 | -0.0349 | No |
| 119 | Slc22a18 | 16690 | -1.152 | 0.0005 | No |