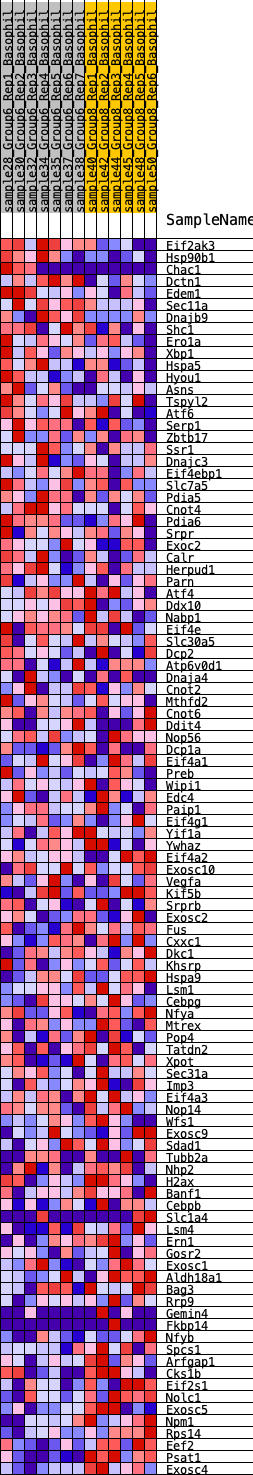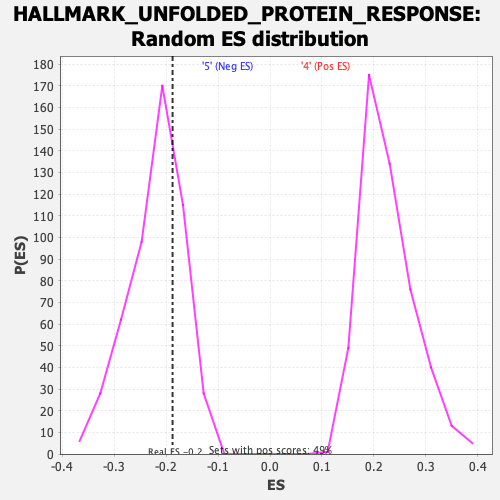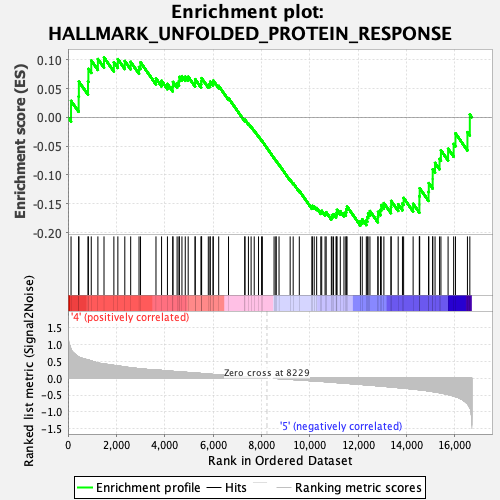
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group6_versus_Group8.Basophil_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | Basophil_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 5 |
| GeneSet | HALLMARK_UNFOLDED_PROTEIN_RESPONSE |
| Enrichment Score (ES) | -0.18779996 |
| Normalized Enrichment Score (NES) | -0.85496634 |
| Nominal p-value | 0.71597636 |
| FDR q-value | 0.82013595 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Eif2ak3 | 126 | 0.860 | 0.0286 | No |
| 2 | Hsp90b1 | 441 | 0.629 | 0.0362 | No |
| 3 | Chac1 | 450 | 0.625 | 0.0620 | No |
| 4 | Dctn1 | 827 | 0.544 | 0.0623 | No |
| 5 | Edem1 | 843 | 0.540 | 0.0841 | No |
| 6 | Sec11a | 962 | 0.514 | 0.0986 | No |
| 7 | Dnajb9 | 1236 | 0.452 | 0.1012 | No |
| 8 | Shc1 | 1489 | 0.427 | 0.1040 | No |
| 9 | Ero1a | 1897 | 0.381 | 0.0955 | No |
| 10 | Xbp1 | 2062 | 0.365 | 0.1010 | No |
| 11 | Hspa5 | 2349 | 0.333 | 0.0978 | No |
| 12 | Hyou1 | 2592 | 0.310 | 0.0963 | No |
| 13 | Asns | 2937 | 0.283 | 0.0875 | No |
| 14 | Tspyl2 | 2999 | 0.278 | 0.0955 | No |
| 15 | Atf6 | 3638 | 0.248 | 0.0675 | No |
| 16 | Serp1 | 3869 | 0.232 | 0.0634 | No |
| 17 | Zbtb17 | 4112 | 0.219 | 0.0580 | No |
| 18 | Ssr1 | 4332 | 0.203 | 0.0534 | No |
| 19 | Dnajc3 | 4341 | 0.203 | 0.0615 | No |
| 20 | Eif4ebp1 | 4512 | 0.198 | 0.0596 | No |
| 21 | Slc7a5 | 4590 | 0.191 | 0.0630 | No |
| 22 | Pdia5 | 4605 | 0.190 | 0.0702 | No |
| 23 | Cnot4 | 4712 | 0.184 | 0.0715 | No |
| 24 | Pdia6 | 4849 | 0.176 | 0.0707 | No |
| 25 | Srpr | 4970 | 0.169 | 0.0706 | No |
| 26 | Exoc2 | 5256 | 0.153 | 0.0599 | No |
| 27 | Calr | 5259 | 0.153 | 0.0662 | No |
| 28 | Herpud1 | 5504 | 0.141 | 0.0575 | No |
| 29 | Parn | 5510 | 0.141 | 0.0631 | No |
| 30 | Atf4 | 5526 | 0.139 | 0.0680 | No |
| 31 | Ddx10 | 5802 | 0.123 | 0.0566 | No |
| 32 | Nabp1 | 5873 | 0.119 | 0.0575 | No |
| 33 | Eif4e | 5881 | 0.119 | 0.0620 | No |
| 34 | Slc30a5 | 5997 | 0.111 | 0.0598 | No |
| 35 | Dcp2 | 6008 | 0.111 | 0.0639 | No |
| 36 | Atp6v0d1 | 6232 | 0.099 | 0.0546 | No |
| 37 | Dnaja4 | 6638 | 0.076 | 0.0334 | No |
| 38 | Cnot2 | 7309 | 0.042 | -0.0052 | No |
| 39 | Mthfd2 | 7319 | 0.042 | -0.0040 | No |
| 40 | Cnot6 | 7461 | 0.036 | -0.0110 | No |
| 41 | Ddit4 | 7572 | 0.031 | -0.0163 | No |
| 42 | Nop56 | 7701 | 0.025 | -0.0229 | No |
| 43 | Dcp1a | 7879 | 0.016 | -0.0329 | No |
| 44 | Eif4a1 | 8005 | 0.009 | -0.0401 | No |
| 45 | Preb | 8037 | 0.007 | -0.0417 | No |
| 46 | Wipi1 | 8522 | -0.001 | -0.0708 | No |
| 47 | Edc4 | 8598 | -0.005 | -0.0751 | No |
| 48 | Paip1 | 8607 | -0.005 | -0.0754 | No |
| 49 | Eif4g1 | 8727 | -0.011 | -0.0821 | No |
| 50 | Yif1a | 9187 | -0.033 | -0.1084 | No |
| 51 | Ywhaz | 9312 | -0.038 | -0.1143 | No |
| 52 | Eif4a2 | 9565 | -0.049 | -0.1274 | No |
| 53 | Exosc10 | 10087 | -0.075 | -0.1556 | No |
| 54 | Vegfa | 10105 | -0.075 | -0.1535 | No |
| 55 | Kif5b | 10182 | -0.078 | -0.1548 | No |
| 56 | Srprb | 10283 | -0.083 | -0.1573 | No |
| 57 | Exosc2 | 10454 | -0.090 | -0.1638 | No |
| 58 | Fus | 10492 | -0.092 | -0.1621 | No |
| 59 | Cxxc1 | 10631 | -0.100 | -0.1662 | No |
| 60 | Dkc1 | 10676 | -0.102 | -0.1646 | No |
| 61 | Khsrp | 10890 | -0.114 | -0.1726 | No |
| 62 | Hspa9 | 10919 | -0.115 | -0.1694 | No |
| 63 | Lsm1 | 10981 | -0.120 | -0.1680 | No |
| 64 | Cebpg | 11084 | -0.125 | -0.1689 | No |
| 65 | Nfya | 11111 | -0.127 | -0.1651 | No |
| 66 | Mtrex | 11123 | -0.128 | -0.1604 | No |
| 67 | Pop4 | 11260 | -0.136 | -0.1629 | No |
| 68 | Tatdn2 | 11412 | -0.143 | -0.1659 | No |
| 69 | Xpot | 11491 | -0.148 | -0.1644 | No |
| 70 | Sec31a | 11507 | -0.149 | -0.1590 | No |
| 71 | Imp3 | 11539 | -0.151 | -0.1545 | No |
| 72 | Eif4a3 | 12092 | -0.181 | -0.1802 | Yes |
| 73 | Nop14 | 12168 | -0.186 | -0.1769 | Yes |
| 74 | Wfs1 | 12336 | -0.195 | -0.1787 | Yes |
| 75 | Exosc9 | 12378 | -0.196 | -0.1729 | Yes |
| 76 | Sdad1 | 12410 | -0.197 | -0.1665 | Yes |
| 77 | Tubb2a | 12486 | -0.202 | -0.1625 | Yes |
| 78 | Nhp2 | 12815 | -0.223 | -0.1729 | Yes |
| 79 | H2ax | 12821 | -0.223 | -0.1638 | Yes |
| 80 | Banf1 | 12924 | -0.230 | -0.1603 | Yes |
| 81 | Cebpb | 12954 | -0.231 | -0.1523 | Yes |
| 82 | Slc1a4 | 13063 | -0.239 | -0.1487 | Yes |
| 83 | Lsm4 | 13356 | -0.258 | -0.1555 | Yes |
| 84 | Ern1 | 13363 | -0.258 | -0.1450 | Yes |
| 85 | Gosr2 | 13654 | -0.278 | -0.1507 | Yes |
| 86 | Exosc1 | 13831 | -0.288 | -0.1492 | Yes |
| 87 | Aldh18a1 | 13879 | -0.291 | -0.1398 | Yes |
| 88 | Bag3 | 14272 | -0.320 | -0.1499 | Yes |
| 89 | Rrp9 | 14531 | -0.341 | -0.1511 | Yes |
| 90 | Gemin4 | 14532 | -0.341 | -0.1368 | Yes |
| 91 | Fkbp14 | 14544 | -0.341 | -0.1230 | Yes |
| 92 | Nfyb | 14910 | -0.375 | -0.1292 | Yes |
| 93 | Spcs1 | 14921 | -0.376 | -0.1140 | Yes |
| 94 | Arfgap1 | 15081 | -0.395 | -0.1070 | Yes |
| 95 | Cks1b | 15082 | -0.395 | -0.0903 | Yes |
| 96 | Eif2s1 | 15179 | -0.406 | -0.0790 | Yes |
| 97 | Nolc1 | 15365 | -0.428 | -0.0721 | Yes |
| 98 | Exosc5 | 15422 | -0.437 | -0.0571 | Yes |
| 99 | Npm1 | 15718 | -0.484 | -0.0544 | Yes |
| 100 | Rps14 | 15948 | -0.527 | -0.0461 | Yes |
| 101 | Eef2 | 16023 | -0.542 | -0.0277 | Yes |
| 102 | Psat1 | 16518 | -0.755 | -0.0257 | Yes |
| 103 | Exosc4 | 16617 | -0.867 | 0.0049 | Yes |