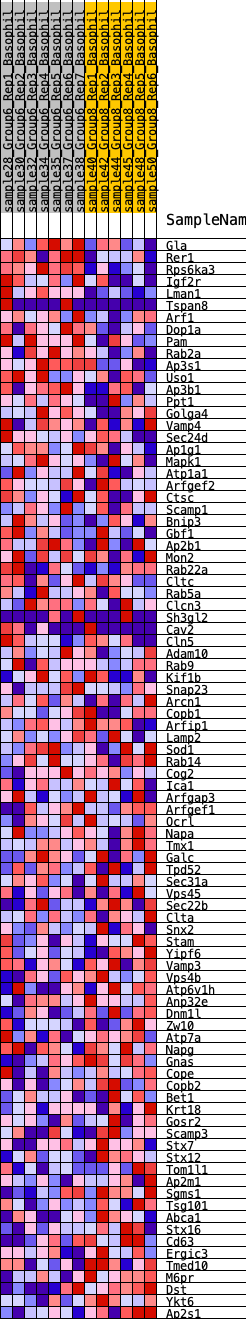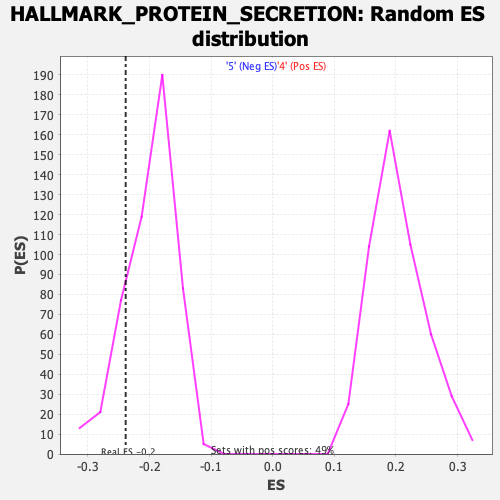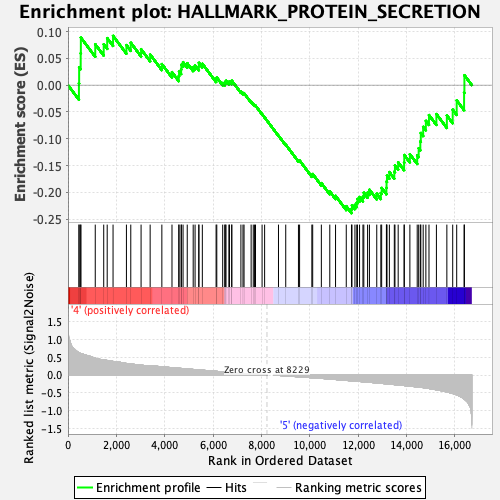
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group6_versus_Group8.Basophil_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | Basophil_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 5 |
| GeneSet | HALLMARK_PROTEIN_SECRETION |
| Enrichment Score (ES) | -0.23878779 |
| Normalized Enrichment Score (NES) | -1.1999843 |
| Nominal p-value | 0.16535433 |
| FDR q-value | 0.5098141 |
| FWER p-Value | 0.958 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Gla | 452 | 0.624 | 0.0034 | No |
| 2 | Rer1 | 461 | 0.621 | 0.0334 | No |
| 3 | Rps6ka3 | 525 | 0.605 | 0.0593 | No |
| 4 | Igf2r | 532 | 0.605 | 0.0886 | No |
| 5 | Lman1 | 1127 | 0.475 | 0.0762 | No |
| 6 | Tspan8 | 1477 | 0.427 | 0.0761 | No |
| 7 | Arf1 | 1621 | 0.410 | 0.0876 | No |
| 8 | Dop1a | 1863 | 0.383 | 0.0919 | No |
| 9 | Pam | 2416 | 0.326 | 0.0746 | No |
| 10 | Rab2a | 2596 | 0.310 | 0.0791 | No |
| 11 | Ap3s1 | 3022 | 0.276 | 0.0670 | No |
| 12 | Uso1 | 3397 | 0.256 | 0.0570 | No |
| 13 | Ap3b1 | 3881 | 0.231 | 0.0393 | No |
| 14 | Ppt1 | 4300 | 0.206 | 0.0242 | No |
| 15 | Golga4 | 4581 | 0.192 | 0.0168 | No |
| 16 | Vamp4 | 4593 | 0.191 | 0.0255 | No |
| 17 | Sec24d | 4678 | 0.186 | 0.0295 | No |
| 18 | Ap1g1 | 4688 | 0.185 | 0.0381 | No |
| 19 | Mapk1 | 4758 | 0.181 | 0.0428 | No |
| 20 | Atp1a1 | 4931 | 0.171 | 0.0408 | No |
| 21 | Arfgef2 | 5177 | 0.157 | 0.0338 | No |
| 22 | Ctsc | 5255 | 0.153 | 0.0367 | No |
| 23 | Scamp1 | 5410 | 0.146 | 0.0346 | No |
| 24 | Bnip3 | 5412 | 0.145 | 0.0417 | No |
| 25 | Gbf1 | 5555 | 0.138 | 0.0398 | No |
| 26 | Ap2b1 | 6124 | 0.105 | 0.0108 | No |
| 27 | Mon2 | 6153 | 0.104 | 0.0142 | No |
| 28 | Rab22a | 6397 | 0.090 | 0.0040 | No |
| 29 | Cltc | 6481 | 0.086 | 0.0032 | No |
| 30 | Rab5a | 6505 | 0.084 | 0.0059 | No |
| 31 | Clcn3 | 6534 | 0.082 | 0.0083 | No |
| 32 | Sh3gl2 | 6654 | 0.075 | 0.0048 | No |
| 33 | Cav2 | 6677 | 0.074 | 0.0071 | No |
| 34 | Cln5 | 6771 | 0.070 | 0.0049 | No |
| 35 | Adam10 | 6773 | 0.069 | 0.0083 | No |
| 36 | Rab9 | 7150 | 0.051 | -0.0119 | No |
| 37 | Kif1b | 7236 | 0.047 | -0.0147 | No |
| 38 | Snap23 | 7280 | 0.045 | -0.0151 | No |
| 39 | Arcn1 | 7573 | 0.031 | -0.0312 | No |
| 40 | Copb1 | 7661 | 0.027 | -0.0351 | No |
| 41 | Arfip1 | 7715 | 0.025 | -0.0371 | No |
| 42 | Lamp2 | 7754 | 0.023 | -0.0382 | No |
| 43 | Sod1 | 8026 | 0.008 | -0.0542 | No |
| 44 | Rab14 | 8131 | 0.003 | -0.0603 | No |
| 45 | Cog2 | 8704 | -0.010 | -0.0942 | No |
| 46 | Ica1 | 9004 | -0.024 | -0.1111 | No |
| 47 | Arfgap3 | 9530 | -0.048 | -0.1404 | No |
| 48 | Arfgef1 | 9577 | -0.050 | -0.1407 | No |
| 49 | Ocrl | 10084 | -0.074 | -0.1675 | No |
| 50 | Napa | 10119 | -0.076 | -0.1658 | No |
| 51 | Tmx1 | 10477 | -0.091 | -0.1828 | No |
| 52 | Galc | 10826 | -0.111 | -0.1983 | No |
| 53 | Tpd52 | 11064 | -0.123 | -0.2065 | No |
| 54 | Sec31a | 11507 | -0.149 | -0.2258 | No |
| 55 | Vps45 | 11723 | -0.161 | -0.2309 | Yes |
| 56 | Sec22b | 11743 | -0.162 | -0.2241 | Yes |
| 57 | Clta | 11866 | -0.168 | -0.2232 | Yes |
| 58 | Snx2 | 11942 | -0.173 | -0.2192 | Yes |
| 59 | Stam | 11970 | -0.174 | -0.2123 | Yes |
| 60 | Yipf6 | 12057 | -0.179 | -0.2087 | Yes |
| 61 | Vamp3 | 12197 | -0.187 | -0.2079 | Yes |
| 62 | Vps4b | 12233 | -0.190 | -0.2007 | Yes |
| 63 | Atp6v1h | 12389 | -0.197 | -0.2004 | Yes |
| 64 | Anp32e | 12465 | -0.200 | -0.1951 | Yes |
| 65 | Dnm1l | 12768 | -0.220 | -0.2025 | Yes |
| 66 | Zw10 | 12937 | -0.231 | -0.2013 | Yes |
| 67 | Atp7a | 12971 | -0.233 | -0.1918 | Yes |
| 68 | Napg | 13167 | -0.245 | -0.1915 | Yes |
| 69 | Gnas | 13173 | -0.246 | -0.1798 | Yes |
| 70 | Cope | 13191 | -0.247 | -0.1687 | Yes |
| 71 | Copb2 | 13290 | -0.253 | -0.1622 | Yes |
| 72 | Bet1 | 13494 | -0.267 | -0.1613 | Yes |
| 73 | Krt18 | 13521 | -0.269 | -0.1497 | Yes |
| 74 | Gosr2 | 13654 | -0.278 | -0.1440 | Yes |
| 75 | Scamp3 | 13899 | -0.293 | -0.1443 | Yes |
| 76 | Stx7 | 13907 | -0.294 | -0.1303 | Yes |
| 77 | Stx12 | 14138 | -0.311 | -0.1289 | Yes |
| 78 | Tom1l1 | 14441 | -0.333 | -0.1308 | Yes |
| 79 | Ap2m1 | 14502 | -0.338 | -0.1178 | Yes |
| 80 | Sgms1 | 14570 | -0.343 | -0.1050 | Yes |
| 81 | Tsg101 | 14590 | -0.344 | -0.0893 | Yes |
| 82 | Abca1 | 14688 | -0.352 | -0.0778 | Yes |
| 83 | Stx16 | 14799 | -0.363 | -0.0666 | Yes |
| 84 | Cd63 | 14928 | -0.377 | -0.0559 | Yes |
| 85 | Ergic3 | 15235 | -0.413 | -0.0540 | Yes |
| 86 | Tmed10 | 15667 | -0.475 | -0.0567 | Yes |
| 87 | M6pr | 15909 | -0.518 | -0.0458 | Yes |
| 88 | Dst | 16074 | -0.554 | -0.0285 | Yes |
| 89 | Ykt6 | 16380 | -0.669 | -0.0140 | Yes |
| 90 | Ap2s1 | 16393 | -0.676 | 0.0184 | Yes |