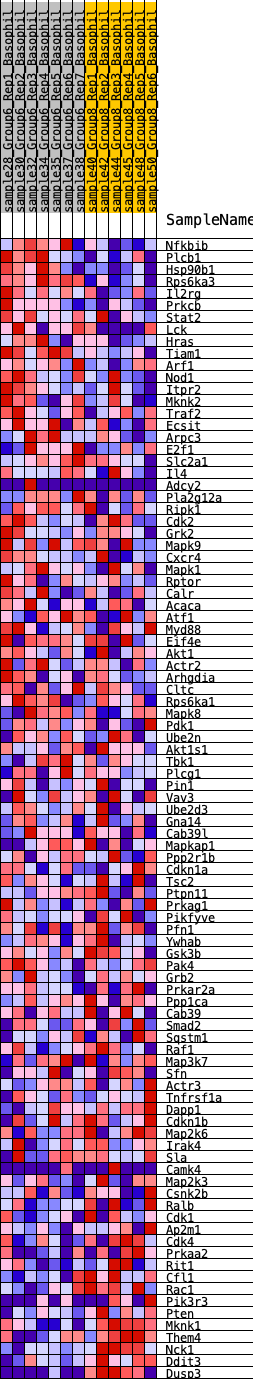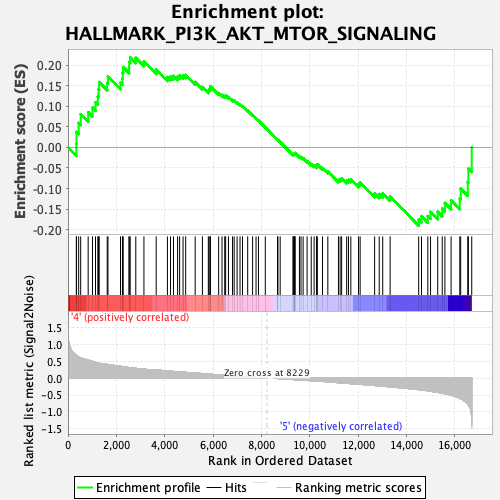
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group6_versus_Group8.Basophil_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | Basophil_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 4 |
| GeneSet | HALLMARK_PI3K_AKT_MTOR_SIGNALING |
| Enrichment Score (ES) | 0.21901791 |
| Normalized Enrichment Score (NES) | 1.0916065 |
| Nominal p-value | 0.27362204 |
| FDR q-value | 0.6334068 |
| FWER p-Value | 0.99 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Nfkbib | 345 | 0.678 | 0.0086 | Yes |
| 2 | Plcb1 | 350 | 0.675 | 0.0377 | Yes |
| 3 | Hsp90b1 | 441 | 0.629 | 0.0596 | Yes |
| 4 | Rps6ka3 | 525 | 0.605 | 0.0809 | Yes |
| 5 | Il2rg | 834 | 0.542 | 0.0858 | Yes |
| 6 | Prkcb | 1013 | 0.499 | 0.0968 | Yes |
| 7 | Stat2 | 1138 | 0.472 | 0.1098 | Yes |
| 8 | Lck | 1234 | 0.452 | 0.1237 | Yes |
| 9 | Hras | 1263 | 0.448 | 0.1415 | Yes |
| 10 | Tiam1 | 1290 | 0.444 | 0.1592 | Yes |
| 11 | Arf1 | 1621 | 0.410 | 0.1571 | Yes |
| 12 | Nod1 | 1653 | 0.407 | 0.1729 | Yes |
| 13 | Itpr2 | 2171 | 0.354 | 0.1571 | Yes |
| 14 | Mknk2 | 2252 | 0.345 | 0.1673 | Yes |
| 15 | Traf2 | 2262 | 0.344 | 0.1817 | Yes |
| 16 | Ecsit | 2280 | 0.342 | 0.1955 | Yes |
| 17 | Arpc3 | 2526 | 0.315 | 0.1944 | Yes |
| 18 | E2f1 | 2528 | 0.315 | 0.2081 | Yes |
| 19 | Slc2a1 | 2572 | 0.312 | 0.2190 | Yes |
| 20 | Il4 | 2803 | 0.293 | 0.2179 | No |
| 21 | Adcy2 | 3140 | 0.268 | 0.2093 | No |
| 22 | Pla2g12a | 3647 | 0.248 | 0.1896 | No |
| 23 | Ripk1 | 4111 | 0.219 | 0.1712 | No |
| 24 | Cdk2 | 4240 | 0.210 | 0.1726 | No |
| 25 | Grk2 | 4362 | 0.201 | 0.1740 | No |
| 26 | Mapk9 | 4532 | 0.196 | 0.1724 | No |
| 27 | Cxcr4 | 4616 | 0.190 | 0.1756 | No |
| 28 | Mapk1 | 4758 | 0.181 | 0.1750 | No |
| 29 | Rptor | 4866 | 0.175 | 0.1761 | No |
| 30 | Calr | 5259 | 0.153 | 0.1591 | No |
| 31 | Acaca | 5558 | 0.137 | 0.1472 | No |
| 32 | Atf1 | 5803 | 0.123 | 0.1378 | No |
| 33 | Myd88 | 5835 | 0.121 | 0.1412 | No |
| 34 | Eif4e | 5881 | 0.119 | 0.1436 | No |
| 35 | Akt1 | 5887 | 0.119 | 0.1485 | No |
| 36 | Actr2 | 6230 | 0.099 | 0.1322 | No |
| 37 | Arhgdia | 6369 | 0.091 | 0.1278 | No |
| 38 | Cltc | 6481 | 0.086 | 0.1249 | No |
| 39 | Rps6ka1 | 6518 | 0.084 | 0.1263 | No |
| 40 | Mapk8 | 6635 | 0.076 | 0.1226 | No |
| 41 | Pdk1 | 6804 | 0.068 | 0.1155 | No |
| 42 | Ube2n | 6872 | 0.065 | 0.1143 | No |
| 43 | Akt1s1 | 6992 | 0.058 | 0.1096 | No |
| 44 | Tbk1 | 7114 | 0.052 | 0.1046 | No |
| 45 | Plcg1 | 7216 | 0.048 | 0.1006 | No |
| 46 | Pin1 | 7431 | 0.037 | 0.0893 | No |
| 47 | Vav3 | 7634 | 0.028 | 0.0784 | No |
| 48 | Ube2d3 | 7776 | 0.022 | 0.0708 | No |
| 49 | Gna14 | 7876 | 0.016 | 0.0656 | No |
| 50 | Cab39l | 8159 | 0.003 | 0.0487 | No |
| 51 | Mapkap1 | 8665 | -0.008 | 0.0186 | No |
| 52 | Ppp2r1b | 8681 | -0.009 | 0.0181 | No |
| 53 | Cdkn1a | 8774 | -0.013 | 0.0131 | No |
| 54 | Tsc2 | 9304 | -0.037 | -0.0171 | No |
| 55 | Ptpn11 | 9319 | -0.038 | -0.0163 | No |
| 56 | Prkag1 | 9350 | -0.039 | -0.0164 | No |
| 57 | Pikfyve | 9362 | -0.040 | -0.0154 | No |
| 58 | Pfn1 | 9379 | -0.041 | -0.0146 | No |
| 59 | Ywhab | 9392 | -0.041 | -0.0135 | No |
| 60 | Gsk3b | 9571 | -0.050 | -0.0221 | No |
| 61 | Pak4 | 9636 | -0.053 | -0.0236 | No |
| 62 | Grb2 | 9723 | -0.058 | -0.0263 | No |
| 63 | Prkar2a | 9887 | -0.065 | -0.0333 | No |
| 64 | Ppp1ca | 10058 | -0.073 | -0.0404 | No |
| 65 | Cab39 | 10178 | -0.078 | -0.0441 | No |
| 66 | Smad2 | 10276 | -0.083 | -0.0464 | No |
| 67 | Sqstm1 | 10282 | -0.083 | -0.0431 | No |
| 68 | Raf1 | 10310 | -0.083 | -0.0411 | No |
| 69 | Map3k7 | 10525 | -0.094 | -0.0499 | No |
| 70 | Sfn | 10745 | -0.106 | -0.0585 | No |
| 71 | Actr3 | 11184 | -0.131 | -0.0792 | No |
| 72 | Tnfrsf1a | 11254 | -0.136 | -0.0774 | No |
| 73 | Dapp1 | 11316 | -0.139 | -0.0750 | No |
| 74 | Cdkn1b | 11524 | -0.150 | -0.0810 | No |
| 75 | Map2k6 | 11599 | -0.155 | -0.0787 | No |
| 76 | Irak4 | 11696 | -0.160 | -0.0776 | No |
| 77 | Sla | 12015 | -0.176 | -0.0891 | No |
| 78 | Camk4 | 12081 | -0.181 | -0.0851 | No |
| 79 | Map2k3 | 12680 | -0.215 | -0.1118 | No |
| 80 | Csnk2b | 12870 | -0.226 | -0.1134 | No |
| 81 | Ralb | 13015 | -0.236 | -0.1118 | No |
| 82 | Cdk1 | 13321 | -0.255 | -0.1191 | No |
| 83 | Ap2m1 | 14502 | -0.338 | -0.1755 | No |
| 84 | Cdk4 | 14618 | -0.347 | -0.1674 | No |
| 85 | Prkaa2 | 14880 | -0.372 | -0.1669 | No |
| 86 | Rit1 | 14992 | -0.384 | -0.1570 | No |
| 87 | Cfl1 | 15289 | -0.418 | -0.1566 | No |
| 88 | Rac1 | 15476 | -0.444 | -0.1486 | No |
| 89 | Pik3r3 | 15586 | -0.461 | -0.1351 | No |
| 90 | Pten | 15842 | -0.504 | -0.1286 | No |
| 91 | Mknk1 | 16202 | -0.593 | -0.1244 | No |
| 92 | Them4 | 16239 | -0.611 | -0.1001 | No |
| 93 | Nck1 | 16532 | -0.764 | -0.0845 | No |
| 94 | Ddit3 | 16557 | -0.795 | -0.0514 | No |
| 95 | Dusp3 | 16696 | -1.380 | 0.0002 | No |