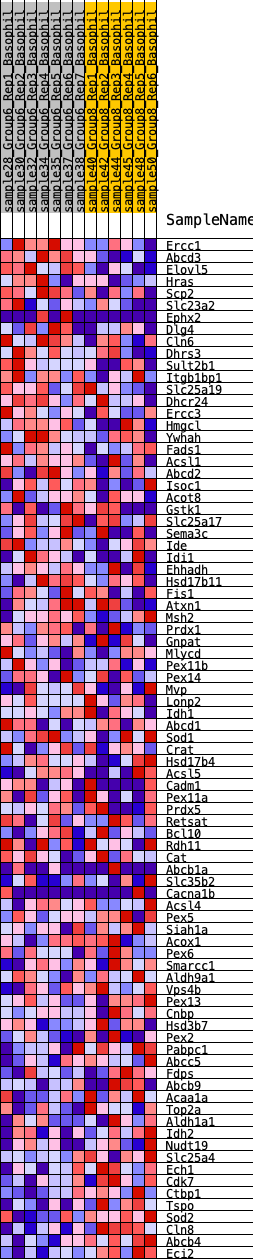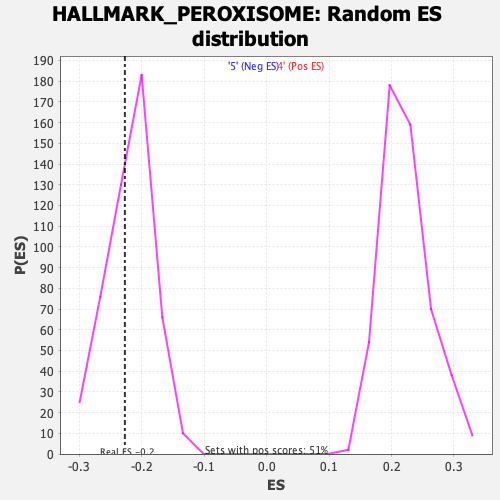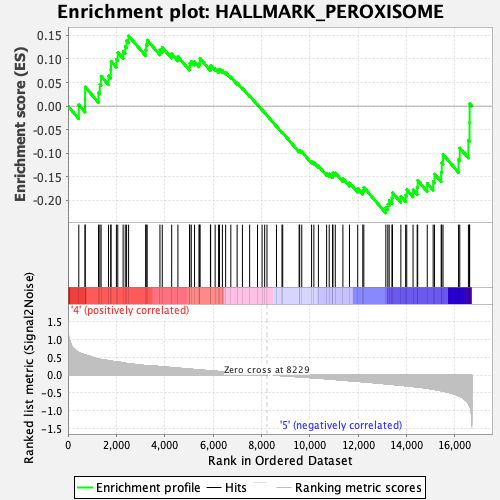
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group6_versus_Group8.Basophil_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | Basophil_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 5 |
| GeneSet | HALLMARK_PEROXISOME |
| Enrichment Score (ES) | -0.2270714 |
| Normalized Enrichment Score (NES) | -1.0414754 |
| Nominal p-value | 0.35714287 |
| FDR q-value | 0.5896408 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ercc1 | 446 | 0.626 | 0.0027 | No |
| 2 | Abcd3 | 706 | 0.568 | 0.0139 | No |
| 3 | Elovl5 | 708 | 0.567 | 0.0406 | No |
| 4 | Hras | 1263 | 0.448 | 0.0284 | No |
| 5 | Scp2 | 1317 | 0.441 | 0.0460 | No |
| 6 | Slc23a2 | 1369 | 0.437 | 0.0635 | No |
| 7 | Ephx2 | 1675 | 0.406 | 0.0643 | No |
| 8 | Dlg4 | 1769 | 0.394 | 0.0773 | No |
| 9 | Cln6 | 1779 | 0.393 | 0.0953 | No |
| 10 | Dhrs3 | 2001 | 0.372 | 0.0996 | No |
| 11 | Sult2b1 | 2058 | 0.366 | 0.1135 | No |
| 12 | Itgb1bp1 | 2279 | 0.342 | 0.1164 | No |
| 13 | Slc25a19 | 2367 | 0.331 | 0.1268 | No |
| 14 | Dhcr24 | 2423 | 0.326 | 0.1388 | No |
| 15 | Ercc3 | 2506 | 0.318 | 0.1489 | No |
| 16 | Hmgcl | 3212 | 0.264 | 0.1190 | No |
| 17 | Ywhah | 3233 | 0.263 | 0.1302 | No |
| 18 | Fads1 | 3274 | 0.260 | 0.1400 | No |
| 19 | Acsl1 | 3802 | 0.236 | 0.1194 | No |
| 20 | Abcd2 | 3898 | 0.230 | 0.1246 | No |
| 21 | Isoc1 | 4288 | 0.207 | 0.1109 | No |
| 22 | Acot8 | 4545 | 0.195 | 0.1047 | No |
| 23 | Gstk1 | 5025 | 0.166 | 0.0837 | No |
| 24 | Slc25a17 | 5036 | 0.165 | 0.0909 | No |
| 25 | Sema3c | 5098 | 0.161 | 0.0948 | No |
| 26 | Ide | 5233 | 0.155 | 0.0941 | No |
| 27 | Idi1 | 5417 | 0.145 | 0.0899 | No |
| 28 | Ehhadh | 5453 | 0.144 | 0.0946 | No |
| 29 | Hsd17b11 | 5454 | 0.144 | 0.1013 | No |
| 30 | Fis1 | 5890 | 0.118 | 0.0807 | No |
| 31 | Atxn1 | 5899 | 0.118 | 0.0858 | No |
| 32 | Msh2 | 6085 | 0.107 | 0.0797 | No |
| 33 | Prdx1 | 6227 | 0.099 | 0.0759 | No |
| 34 | Gnpat | 6266 | 0.097 | 0.0782 | No |
| 35 | Mlycd | 6384 | 0.090 | 0.0754 | No |
| 36 | Pex11b | 6519 | 0.083 | 0.0713 | No |
| 37 | Pex14 | 6733 | 0.071 | 0.0618 | No |
| 38 | Mvp | 6994 | 0.058 | 0.0490 | No |
| 39 | Lonp2 | 7213 | 0.048 | 0.0381 | No |
| 40 | Idh1 | 7512 | 0.033 | 0.0217 | No |
| 41 | Abcd1 | 7837 | 0.018 | 0.0031 | No |
| 42 | Sod1 | 8026 | 0.008 | -0.0079 | No |
| 43 | Crat | 8132 | 0.003 | -0.0140 | No |
| 44 | Hsd17b4 | 8228 | 0.000 | -0.0198 | No |
| 45 | Acsl5 | 8621 | -0.005 | -0.0431 | No |
| 46 | Cadm1 | 8849 | -0.016 | -0.0560 | No |
| 47 | Pex11a | 8880 | -0.018 | -0.0569 | No |
| 48 | Prdx5 | 9558 | -0.049 | -0.0954 | No |
| 49 | Retsat | 9581 | -0.050 | -0.0943 | No |
| 50 | Bcl10 | 9667 | -0.054 | -0.0969 | No |
| 51 | Rdh11 | 10073 | -0.074 | -0.1178 | No |
| 52 | Cat | 10168 | -0.078 | -0.1198 | No |
| 53 | Abcb1a | 10355 | -0.086 | -0.1269 | No |
| 54 | Slc35b2 | 10695 | -0.103 | -0.1424 | No |
| 55 | Cacna1b | 10802 | -0.110 | -0.1436 | No |
| 56 | Acsl4 | 10941 | -0.117 | -0.1464 | No |
| 57 | Pex5 | 10955 | -0.118 | -0.1416 | No |
| 58 | Siah1a | 11053 | -0.123 | -0.1417 | No |
| 59 | Acox1 | 11368 | -0.142 | -0.1539 | No |
| 60 | Pex6 | 11640 | -0.156 | -0.1628 | No |
| 61 | Smarcc1 | 11980 | -0.175 | -0.1750 | No |
| 62 | Aldh9a1 | 12191 | -0.187 | -0.1788 | No |
| 63 | Vps4b | 12233 | -0.190 | -0.1723 | No |
| 64 | Pex13 | 13144 | -0.243 | -0.2156 | Yes |
| 65 | Cnbp | 13225 | -0.249 | -0.2086 | Yes |
| 66 | Hsd3b7 | 13281 | -0.252 | -0.2001 | Yes |
| 67 | Pex2 | 13396 | -0.261 | -0.1946 | Yes |
| 68 | Pabpc1 | 13419 | -0.262 | -0.1836 | Yes |
| 69 | Abcc5 | 13765 | -0.282 | -0.1911 | Yes |
| 70 | Fdps | 13963 | -0.299 | -0.1888 | Yes |
| 71 | Abcb9 | 14008 | -0.303 | -0.1772 | Yes |
| 72 | Acaa1a | 14269 | -0.320 | -0.1777 | Yes |
| 73 | Top2a | 14440 | -0.333 | -0.1722 | Yes |
| 74 | Aldh1a1 | 14461 | -0.335 | -0.1576 | Yes |
| 75 | Idh2 | 14856 | -0.370 | -0.1639 | Yes |
| 76 | Nudt19 | 15100 | -0.397 | -0.1598 | Yes |
| 77 | Slc25a4 | 15157 | -0.404 | -0.1441 | Yes |
| 78 | Ech1 | 15432 | -0.438 | -0.1399 | Yes |
| 79 | Cdk7 | 15455 | -0.441 | -0.1204 | Yes |
| 80 | Ctbp1 | 15509 | -0.448 | -0.1025 | Yes |
| 81 | Tspo | 16151 | -0.577 | -0.1138 | Yes |
| 82 | Sod2 | 16201 | -0.593 | -0.0888 | Yes |
| 83 | Cln8 | 16559 | -0.800 | -0.0725 | Yes |
| 84 | Abcb4 | 16605 | -0.854 | -0.0349 | Yes |
| 85 | Eci2 | 16609 | -0.858 | 0.0054 | Yes |