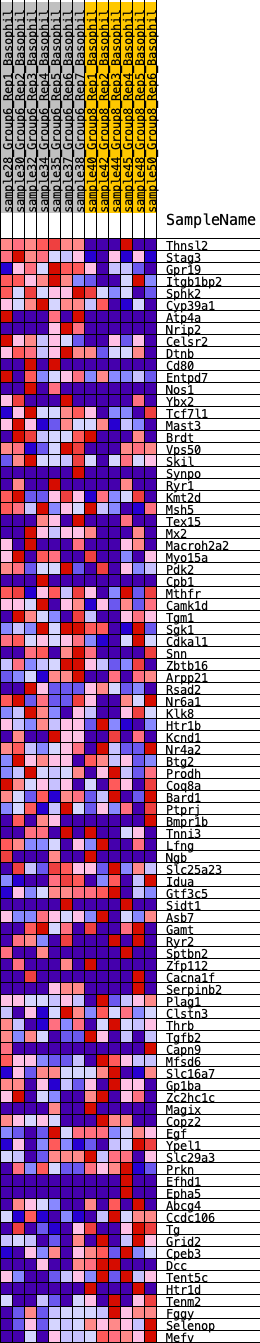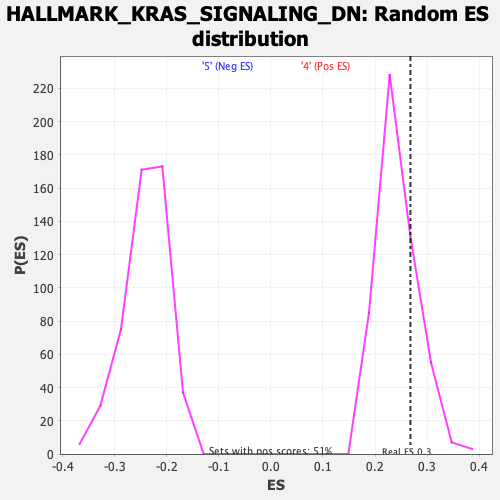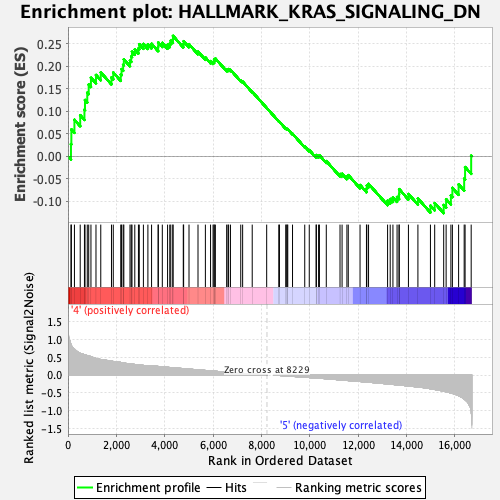
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group6_versus_Group8.Basophil_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | Basophil_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 4 |
| GeneSet | HALLMARK_KRAS_SIGNALING_DN |
| Enrichment Score (ES) | 0.2679791 |
| Normalized Enrichment Score (NES) | 1.1012561 |
| Nominal p-value | 0.25540274 |
| FDR q-value | 0.6527795 |
| FWER p-Value | 0.99 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Thnsl2 | 123 | 0.863 | 0.0272 | Yes |
| 2 | Stag3 | 139 | 0.835 | 0.0597 | Yes |
| 3 | Gpr19 | 266 | 0.728 | 0.0813 | Yes |
| 4 | Itgb1bp2 | 505 | 0.609 | 0.0913 | Yes |
| 5 | Sphk2 | 680 | 0.574 | 0.1038 | Yes |
| 6 | Cyp39a1 | 703 | 0.568 | 0.1253 | Yes |
| 7 | Atp4a | 797 | 0.549 | 0.1417 | Yes |
| 8 | Nrip2 | 856 | 0.536 | 0.1596 | Yes |
| 9 | Celsr2 | 949 | 0.516 | 0.1747 | Yes |
| 10 | Dtnb | 1157 | 0.466 | 0.1809 | Yes |
| 11 | Cd80 | 1355 | 0.438 | 0.1866 | Yes |
| 12 | Entpd7 | 1798 | 0.389 | 0.1756 | Yes |
| 13 | Nos1 | 1872 | 0.381 | 0.1864 | Yes |
| 14 | Ybx2 | 2182 | 0.353 | 0.1820 | Yes |
| 15 | Tcf7l1 | 2215 | 0.349 | 0.1940 | Yes |
| 16 | Mast3 | 2281 | 0.342 | 0.2038 | Yes |
| 17 | Brdt | 2309 | 0.339 | 0.2157 | Yes |
| 18 | Vps50 | 2564 | 0.313 | 0.2129 | Yes |
| 19 | Skil | 2615 | 0.308 | 0.2223 | Yes |
| 20 | Synpo | 2646 | 0.307 | 0.2327 | Yes |
| 21 | Ryr1 | 2762 | 0.297 | 0.2377 | Yes |
| 22 | Kmt2d | 2913 | 0.285 | 0.2401 | Yes |
| 23 | Msh5 | 2947 | 0.281 | 0.2494 | Yes |
| 24 | Tex15 | 3118 | 0.271 | 0.2500 | Yes |
| 25 | Mx2 | 3304 | 0.259 | 0.2492 | Yes |
| 26 | Macroh2a2 | 3456 | 0.256 | 0.2504 | Yes |
| 27 | Myo15a | 3729 | 0.242 | 0.2437 | Yes |
| 28 | Pdk2 | 3732 | 0.242 | 0.2532 | Yes |
| 29 | Cpb1 | 3900 | 0.230 | 0.2524 | Yes |
| 30 | Mthfr | 4118 | 0.218 | 0.2481 | Yes |
| 31 | Camk1d | 4209 | 0.213 | 0.2512 | Yes |
| 32 | Tgm1 | 4250 | 0.209 | 0.2572 | Yes |
| 33 | Sgk1 | 4337 | 0.203 | 0.2601 | Yes |
| 34 | Cdkal1 | 4342 | 0.203 | 0.2680 | Yes |
| 35 | Snn | 4767 | 0.180 | 0.2497 | No |
| 36 | Zbtb16 | 4782 | 0.180 | 0.2560 | No |
| 37 | Arpp21 | 5004 | 0.167 | 0.2494 | No |
| 38 | Rsad2 | 5376 | 0.148 | 0.2330 | No |
| 39 | Nr6a1 | 5681 | 0.129 | 0.2198 | No |
| 40 | Klk8 | 5895 | 0.118 | 0.2117 | No |
| 41 | Htr1b | 5999 | 0.111 | 0.2100 | No |
| 42 | Kcnd1 | 6043 | 0.109 | 0.2118 | No |
| 43 | Nr4a2 | 6044 | 0.109 | 0.2161 | No |
| 44 | Btg2 | 6094 | 0.107 | 0.2175 | No |
| 45 | Prodh | 6561 | 0.081 | 0.1926 | No |
| 46 | Coq8a | 6616 | 0.077 | 0.1925 | No |
| 47 | Bard1 | 6642 | 0.076 | 0.1940 | No |
| 48 | Ptprj | 6718 | 0.072 | 0.1924 | No |
| 49 | Bmpr1b | 7144 | 0.051 | 0.1688 | No |
| 50 | Tnni3 | 7222 | 0.048 | 0.1661 | No |
| 51 | Lfng | 7618 | 0.029 | 0.1435 | No |
| 52 | Ngb | 8216 | 0.000 | 0.1075 | No |
| 53 | Slc25a23 | 8717 | -0.010 | 0.0779 | No |
| 54 | Idua | 8744 | -0.011 | 0.0767 | No |
| 55 | Gtf3c5 | 9007 | -0.024 | 0.0619 | No |
| 56 | Sidt1 | 9025 | -0.025 | 0.0619 | No |
| 57 | Asb7 | 9065 | -0.027 | 0.0606 | No |
| 58 | Gamt | 9088 | -0.028 | 0.0604 | No |
| 59 | Ryr2 | 9285 | -0.037 | 0.0501 | No |
| 60 | Sptbn2 | 9790 | -0.060 | 0.0221 | No |
| 61 | Zfp112 | 9977 | -0.069 | 0.0137 | No |
| 62 | Cacna1f | 10260 | -0.082 | -0.0000 | No |
| 63 | Serpinb2 | 10271 | -0.082 | 0.0027 | No |
| 64 | Plag1 | 10363 | -0.086 | 0.0007 | No |
| 65 | Clstn3 | 10397 | -0.087 | 0.0022 | No |
| 66 | Thrb | 10681 | -0.103 | -0.0108 | No |
| 67 | Tgfb2 | 11249 | -0.136 | -0.0395 | No |
| 68 | Capn9 | 11334 | -0.140 | -0.0389 | No |
| 69 | Mfsd6 | 11534 | -0.151 | -0.0449 | No |
| 70 | Slc16a7 | 11590 | -0.154 | -0.0420 | No |
| 71 | Gp1ba | 12077 | -0.181 | -0.0641 | No |
| 72 | Zc2hc1c | 12342 | -0.195 | -0.0722 | No |
| 73 | Magix | 12358 | -0.195 | -0.0652 | No |
| 74 | Copz2 | 12429 | -0.198 | -0.0615 | No |
| 75 | Egf | 13216 | -0.248 | -0.0989 | No |
| 76 | Ypel1 | 13324 | -0.255 | -0.0951 | No |
| 77 | Slc29a3 | 13437 | -0.263 | -0.0913 | No |
| 78 | Prkn | 13608 | -0.275 | -0.0906 | No |
| 79 | Efhd1 | 13692 | -0.280 | -0.0843 | No |
| 80 | Epha5 | 13695 | -0.280 | -0.0732 | No |
| 81 | Abcg4 | 14078 | -0.305 | -0.0840 | No |
| 82 | Ccdc106 | 14470 | -0.336 | -0.0941 | No |
| 83 | Tg | 14988 | -0.383 | -0.1099 | No |
| 84 | Grid2 | 15164 | -0.404 | -0.1042 | No |
| 85 | Cpeb3 | 15536 | -0.452 | -0.1085 | No |
| 86 | Dcc | 15637 | -0.470 | -0.0957 | No |
| 87 | Tent5c | 15834 | -0.503 | -0.0873 | No |
| 88 | Htr1d | 15896 | -0.516 | -0.0704 | No |
| 89 | Tenm2 | 16153 | -0.578 | -0.0626 | No |
| 90 | Fggy | 16383 | -0.670 | -0.0496 | No |
| 91 | Selenop | 16421 | -0.693 | -0.0241 | No |
| 92 | Mefv | 16673 | -1.018 | 0.0016 | No |