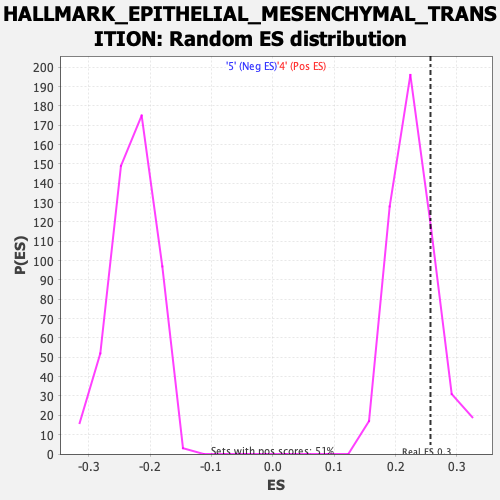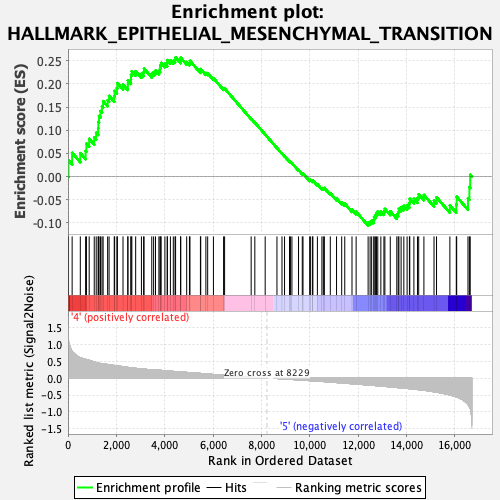
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group6_versus_Group8.Basophil_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | Basophil_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 4 |
| GeneSet | HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION |
| Enrichment Score (ES) | 0.25692767 |
| Normalized Enrichment Score (NES) | 1.1228508 |
| Nominal p-value | 0.17913386 |
| FDR q-value | 0.63563263 |
| FWER p-Value | 0.986 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Sntb1 | 22 | 1.126 | 0.0349 | Yes |
| 2 | Cald1 | 171 | 0.788 | 0.0513 | Yes |
| 3 | Comp | 509 | 0.608 | 0.0505 | Yes |
| 4 | Serpine2 | 729 | 0.561 | 0.0553 | Yes |
| 5 | Il15 | 763 | 0.553 | 0.0711 | Yes |
| 6 | Gadd45a | 876 | 0.533 | 0.0815 | Yes |
| 7 | Tpm1 | 1085 | 0.483 | 0.0845 | Yes |
| 8 | Pcolce | 1166 | 0.464 | 0.0946 | Yes |
| 9 | Mmp2 | 1249 | 0.450 | 0.1041 | Yes |
| 10 | Nt5e | 1255 | 0.448 | 0.1182 | Yes |
| 11 | Sat1 | 1284 | 0.445 | 0.1308 | Yes |
| 12 | Adam12 | 1345 | 0.438 | 0.1413 | Yes |
| 13 | Fgf2 | 1406 | 0.434 | 0.1516 | Yes |
| 14 | Fstl3 | 1452 | 0.429 | 0.1627 | Yes |
| 15 | Lama2 | 1639 | 0.408 | 0.1646 | Yes |
| 16 | Lamc1 | 1699 | 0.405 | 0.1740 | Yes |
| 17 | Bgn | 1912 | 0.378 | 0.1734 | Yes |
| 18 | Gem | 1929 | 0.378 | 0.1846 | Yes |
| 19 | Tpm2 | 2022 | 0.369 | 0.1909 | Yes |
| 20 | Il6 | 2043 | 0.367 | 0.2015 | Yes |
| 21 | Edil3 | 2275 | 0.342 | 0.1986 | Yes |
| 22 | Lrp1 | 2475 | 0.321 | 0.1969 | Yes |
| 23 | Plod1 | 2476 | 0.321 | 0.2073 | Yes |
| 24 | Calu | 2603 | 0.309 | 0.2096 | Yes |
| 25 | Itga2 | 2606 | 0.309 | 0.2194 | Yes |
| 26 | Id2 | 2640 | 0.307 | 0.2273 | Yes |
| 27 | Wipf1 | 2798 | 0.294 | 0.2273 | Yes |
| 28 | Lama3 | 3036 | 0.275 | 0.2218 | Yes |
| 29 | Glipr1 | 3128 | 0.269 | 0.2250 | Yes |
| 30 | Efemp2 | 3144 | 0.268 | 0.2327 | Yes |
| 31 | Dab2 | 3464 | 0.255 | 0.2217 | Yes |
| 32 | Flna | 3541 | 0.254 | 0.2252 | Yes |
| 33 | Fas | 3619 | 0.250 | 0.2286 | Yes |
| 34 | Thbs1 | 3759 | 0.240 | 0.2280 | Yes |
| 35 | Notch2 | 3810 | 0.236 | 0.2325 | Yes |
| 36 | Magee1 | 3816 | 0.235 | 0.2398 | Yes |
| 37 | Copa | 3852 | 0.233 | 0.2452 | Yes |
| 38 | Col4a2 | 4007 | 0.227 | 0.2432 | Yes |
| 39 | Ecm2 | 4103 | 0.219 | 0.2445 | Yes |
| 40 | Tnfaip3 | 4109 | 0.219 | 0.2512 | Yes |
| 41 | Fuca1 | 4235 | 0.211 | 0.2505 | Yes |
| 42 | Fap | 4355 | 0.202 | 0.2498 | Yes |
| 43 | Pfn2 | 4426 | 0.198 | 0.2519 | Yes |
| 44 | Mest | 4449 | 0.198 | 0.2569 | Yes |
| 45 | Anpep | 4656 | 0.187 | 0.2505 | No |
| 46 | Jun | 4662 | 0.187 | 0.2562 | No |
| 47 | Capg | 4908 | 0.173 | 0.2470 | No |
| 48 | Tgm2 | 5023 | 0.166 | 0.2455 | No |
| 49 | Cap2 | 5044 | 0.164 | 0.2495 | No |
| 50 | Itgb3 | 5476 | 0.142 | 0.2281 | No |
| 51 | Qsox1 | 5489 | 0.142 | 0.2320 | No |
| 52 | Bmp1 | 5702 | 0.128 | 0.2233 | No |
| 53 | Cdh11 | 5776 | 0.124 | 0.2229 | No |
| 54 | Itgav | 6014 | 0.110 | 0.2121 | No |
| 55 | Lgals1 | 6434 | 0.088 | 0.1897 | No |
| 56 | Cdh2 | 6476 | 0.086 | 0.1900 | No |
| 57 | Nid2 | 7574 | 0.031 | 0.1249 | No |
| 58 | Gja1 | 7726 | 0.024 | 0.1165 | No |
| 59 | Vegfc | 8154 | 0.003 | 0.0909 | No |
| 60 | Cd44 | 8641 | -0.006 | 0.0618 | No |
| 61 | Cadm1 | 8849 | -0.016 | 0.0498 | No |
| 62 | Tgfbr3 | 8955 | -0.022 | 0.0442 | No |
| 63 | Tgfbi | 9160 | -0.031 | 0.0329 | No |
| 64 | Thy1 | 9198 | -0.033 | 0.0317 | No |
| 65 | Mcm7 | 9260 | -0.036 | 0.0292 | No |
| 66 | Slit3 | 9533 | -0.048 | 0.0143 | No |
| 67 | Spp1 | 9688 | -0.056 | 0.0068 | No |
| 68 | Col4a1 | 9718 | -0.057 | 0.0069 | No |
| 69 | Plod2 | 9995 | -0.070 | -0.0075 | No |
| 70 | Timp3 | 10023 | -0.072 | -0.0068 | No |
| 71 | Vegfa | 10105 | -0.075 | -0.0093 | No |
| 72 | Tpm4 | 10127 | -0.076 | -0.0081 | No |
| 73 | Col16a1 | 10313 | -0.084 | -0.0166 | No |
| 74 | Rhob | 10500 | -0.093 | -0.0248 | No |
| 75 | Tgfb1 | 10560 | -0.096 | -0.0253 | No |
| 76 | Itga5 | 10602 | -0.099 | -0.0246 | No |
| 77 | Tnfrsf12a | 10847 | -0.112 | -0.0357 | No |
| 78 | Col5a1 | 11105 | -0.127 | -0.0471 | No |
| 79 | Vim | 11318 | -0.139 | -0.0554 | No |
| 80 | P3h1 | 11445 | -0.145 | -0.0583 | No |
| 81 | Ntm | 11744 | -0.162 | -0.0711 | No |
| 82 | Itgb1 | 11915 | -0.171 | -0.0758 | No |
| 83 | Itgb5 | 12411 | -0.197 | -0.0993 | No |
| 84 | Fbn1 | 12493 | -0.203 | -0.0977 | No |
| 85 | Plod3 | 12557 | -0.207 | -0.0948 | No |
| 86 | Col7a1 | 12648 | -0.213 | -0.0934 | No |
| 87 | Ecm1 | 12657 | -0.214 | -0.0870 | No |
| 88 | Fn1 | 12712 | -0.217 | -0.0833 | No |
| 89 | Vcan | 12756 | -0.219 | -0.0788 | No |
| 90 | Ppib | 12810 | -0.222 | -0.0749 | No |
| 91 | Sgcd | 12938 | -0.231 | -0.0751 | No |
| 92 | Col1a2 | 13064 | -0.239 | -0.0750 | No |
| 93 | Matn2 | 13104 | -0.241 | -0.0696 | No |
| 94 | Pvr | 13327 | -0.255 | -0.0748 | No |
| 95 | Colgalt1 | 13597 | -0.274 | -0.0822 | No |
| 96 | Emp3 | 13656 | -0.278 | -0.0767 | No |
| 97 | Slc6a8 | 13684 | -0.280 | -0.0694 | No |
| 98 | Gpc1 | 13775 | -0.284 | -0.0657 | No |
| 99 | Col1a1 | 13888 | -0.292 | -0.0630 | No |
| 100 | Gadd45b | 14023 | -0.304 | -0.0613 | No |
| 101 | Vcam1 | 14119 | -0.309 | -0.0571 | No |
| 102 | Basp1 | 14134 | -0.311 | -0.0480 | No |
| 103 | Mmp14 | 14304 | -0.322 | -0.0478 | No |
| 104 | Pmepa1 | 14452 | -0.334 | -0.0459 | No |
| 105 | Fstl1 | 14503 | -0.338 | -0.0381 | No |
| 106 | Slit2 | 14718 | -0.355 | -0.0395 | No |
| 107 | Plaur | 15136 | -0.402 | -0.0518 | No |
| 108 | Sfrp4 | 15240 | -0.413 | -0.0447 | No |
| 109 | Mylk | 15792 | -0.494 | -0.0620 | No |
| 110 | Fzd8 | 16057 | -0.552 | -0.0602 | No |
| 111 | Dst | 16074 | -0.554 | -0.0434 | No |
| 112 | Sgcb | 16541 | -0.774 | -0.0466 | No |
| 113 | Igfbp4 | 16599 | -0.848 | -0.0228 | No |
| 114 | Pdgfrb | 16633 | -0.894 | 0.0040 | No |