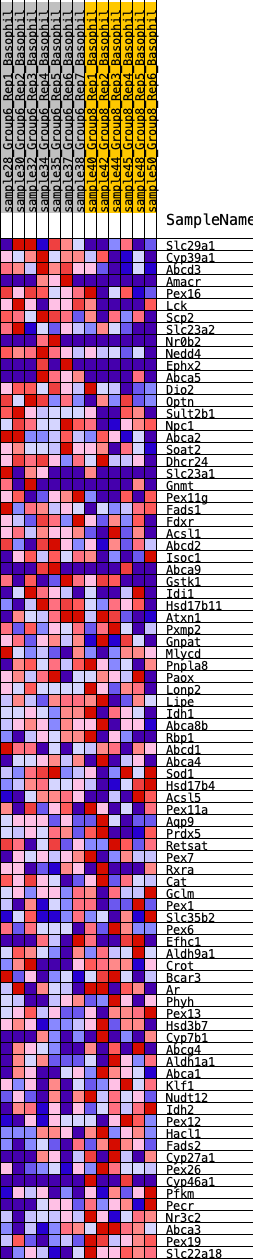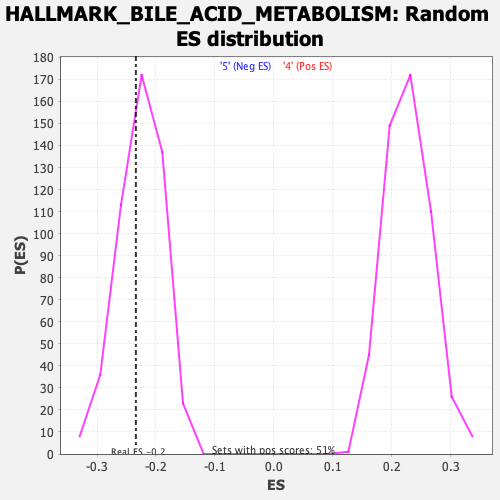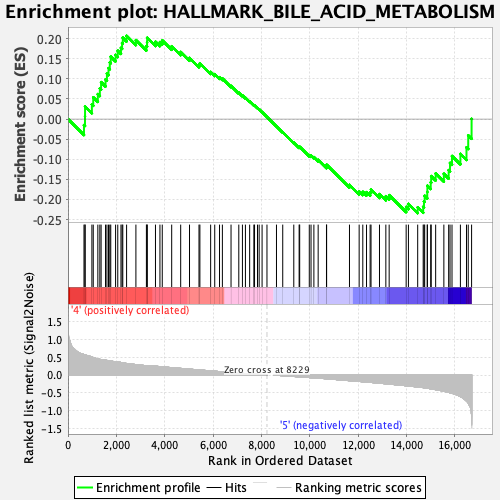
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group6_versus_Group8.Basophil_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | Basophil_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 5 |
| GeneSet | HALLMARK_BILE_ACID_METABOLISM |
| Enrichment Score (ES) | -0.23410733 |
| Normalized Enrichment Score (NES) | -1.034185 |
| Nominal p-value | 0.398773 |
| FDR q-value | 0.5676927 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Slc29a1 | 661 | 0.578 | -0.0149 | No |
| 2 | Cyp39a1 | 703 | 0.568 | 0.0071 | No |
| 3 | Abcd3 | 706 | 0.568 | 0.0314 | No |
| 4 | Amacr | 988 | 0.506 | 0.0362 | No |
| 5 | Pex16 | 1044 | 0.492 | 0.0541 | No |
| 6 | Lck | 1234 | 0.452 | 0.0622 | No |
| 7 | Scp2 | 1317 | 0.441 | 0.0762 | No |
| 8 | Slc23a2 | 1369 | 0.437 | 0.0919 | No |
| 9 | Nr0b2 | 1552 | 0.417 | 0.0989 | No |
| 10 | Nedd4 | 1610 | 0.411 | 0.1131 | No |
| 11 | Ephx2 | 1675 | 0.406 | 0.1267 | No |
| 12 | Abca5 | 1729 | 0.400 | 0.1408 | No |
| 13 | Dio2 | 1768 | 0.394 | 0.1554 | No |
| 14 | Optn | 1968 | 0.376 | 0.1597 | No |
| 15 | Sult2b1 | 2058 | 0.366 | 0.1700 | No |
| 16 | Npc1 | 2190 | 0.352 | 0.1773 | No |
| 17 | Abca2 | 2237 | 0.346 | 0.1894 | No |
| 18 | Soat2 | 2266 | 0.343 | 0.2025 | No |
| 19 | Dhcr24 | 2423 | 0.326 | 0.2071 | No |
| 20 | Slc23a1 | 2808 | 0.293 | 0.1966 | No |
| 21 | Gnmt | 3242 | 0.262 | 0.1818 | No |
| 22 | Pex11g | 3270 | 0.260 | 0.1914 | No |
| 23 | Fads1 | 3274 | 0.260 | 0.2024 | No |
| 24 | Fdxr | 3617 | 0.250 | 0.1925 | No |
| 25 | Acsl1 | 3802 | 0.236 | 0.1916 | No |
| 26 | Abcd2 | 3898 | 0.230 | 0.1958 | No |
| 27 | Isoc1 | 4288 | 0.207 | 0.1813 | No |
| 28 | Abca9 | 4660 | 0.187 | 0.1670 | No |
| 29 | Gstk1 | 5025 | 0.166 | 0.1522 | No |
| 30 | Idi1 | 5417 | 0.145 | 0.1349 | No |
| 31 | Hsd17b11 | 5454 | 0.144 | 0.1389 | No |
| 32 | Atxn1 | 5899 | 0.118 | 0.1173 | No |
| 33 | Pxmp2 | 6068 | 0.108 | 0.1118 | No |
| 34 | Gnpat | 6266 | 0.097 | 0.1041 | No |
| 35 | Mlycd | 6384 | 0.090 | 0.1010 | No |
| 36 | Pnpla8 | 6742 | 0.071 | 0.0825 | No |
| 37 | Paox | 7063 | 0.055 | 0.0656 | No |
| 38 | Lonp2 | 7213 | 0.048 | 0.0587 | No |
| 39 | Lipe | 7337 | 0.041 | 0.0531 | No |
| 40 | Idh1 | 7512 | 0.033 | 0.0441 | No |
| 41 | Abca8b | 7686 | 0.026 | 0.0348 | No |
| 42 | Rbp1 | 7714 | 0.025 | 0.0342 | No |
| 43 | Abcd1 | 7837 | 0.018 | 0.0276 | No |
| 44 | Abca4 | 7905 | 0.015 | 0.0242 | No |
| 45 | Sod1 | 8026 | 0.008 | 0.0173 | No |
| 46 | Hsd17b4 | 8228 | 0.000 | 0.0052 | No |
| 47 | Acsl5 | 8621 | -0.005 | -0.0181 | No |
| 48 | Pex11a | 8880 | -0.018 | -0.0329 | No |
| 49 | Aqp9 | 9337 | -0.039 | -0.0587 | No |
| 50 | Prdx5 | 9558 | -0.049 | -0.0698 | No |
| 51 | Retsat | 9581 | -0.050 | -0.0690 | No |
| 52 | Pex7 | 9978 | -0.069 | -0.0898 | No |
| 53 | Rxra | 10050 | -0.073 | -0.0910 | No |
| 54 | Cat | 10168 | -0.078 | -0.0947 | No |
| 55 | Gclm | 10345 | -0.085 | -0.1016 | No |
| 56 | Pex1 | 10689 | -0.103 | -0.1178 | No |
| 57 | Slc35b2 | 10695 | -0.103 | -0.1137 | No |
| 58 | Pex6 | 11640 | -0.156 | -0.1638 | No |
| 59 | Efhc1 | 12040 | -0.178 | -0.1801 | No |
| 60 | Aldh9a1 | 12191 | -0.187 | -0.1811 | No |
| 61 | Crot | 12345 | -0.195 | -0.1819 | No |
| 62 | Bcar3 | 12494 | -0.203 | -0.1821 | No |
| 63 | Ar | 12531 | -0.205 | -0.1754 | No |
| 64 | Phyh | 12881 | -0.227 | -0.1867 | No |
| 65 | Pex13 | 13144 | -0.243 | -0.1920 | No |
| 66 | Hsd3b7 | 13281 | -0.252 | -0.1893 | No |
| 67 | Cyp7b1 | 13984 | -0.301 | -0.2186 | No |
| 68 | Abcg4 | 14078 | -0.305 | -0.2111 | No |
| 69 | Aldh1a1 | 14461 | -0.335 | -0.2197 | Yes |
| 70 | Abca1 | 14688 | -0.352 | -0.2182 | Yes |
| 71 | Klf1 | 14721 | -0.356 | -0.2048 | Yes |
| 72 | Nudt12 | 14750 | -0.359 | -0.1910 | Yes |
| 73 | Idh2 | 14856 | -0.370 | -0.1814 | Yes |
| 74 | Pex12 | 14862 | -0.371 | -0.1658 | Yes |
| 75 | Hacl1 | 15000 | -0.386 | -0.1575 | Yes |
| 76 | Fads2 | 15019 | -0.388 | -0.1419 | Yes |
| 77 | Cyp27a1 | 15204 | -0.409 | -0.1353 | Yes |
| 78 | Pex26 | 15543 | -0.454 | -0.1362 | Yes |
| 79 | Cyp46a1 | 15742 | -0.488 | -0.1271 | Yes |
| 80 | Pfkm | 15798 | -0.496 | -0.1091 | Yes |
| 81 | Pecr | 15880 | -0.513 | -0.0919 | Yes |
| 82 | Nr3c2 | 16227 | -0.605 | -0.0867 | Yes |
| 83 | Abca3 | 16479 | -0.732 | -0.0703 | Yes |
| 84 | Pex19 | 16551 | -0.788 | -0.0407 | Yes |
| 85 | Slc22a18 | 16690 | -1.152 | 0.0005 | Yes |