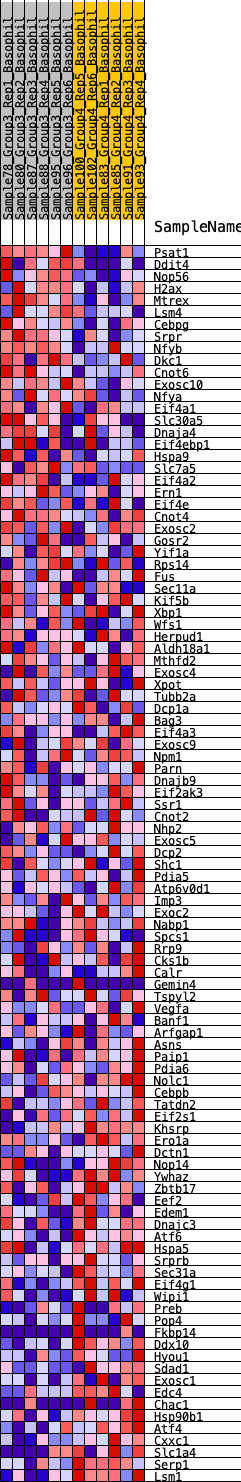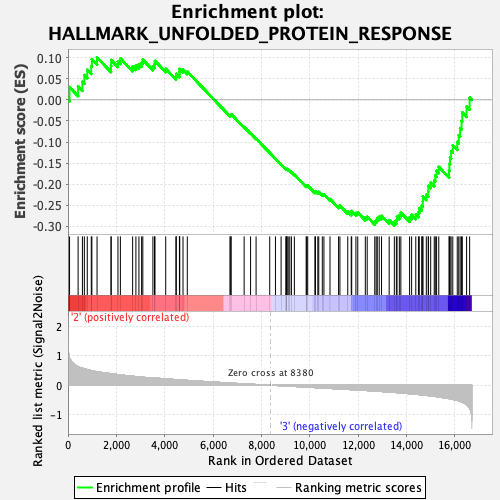
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group3_versus_Group4.Basophil_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_UNFOLDED_PROTEIN_RESPONSE |
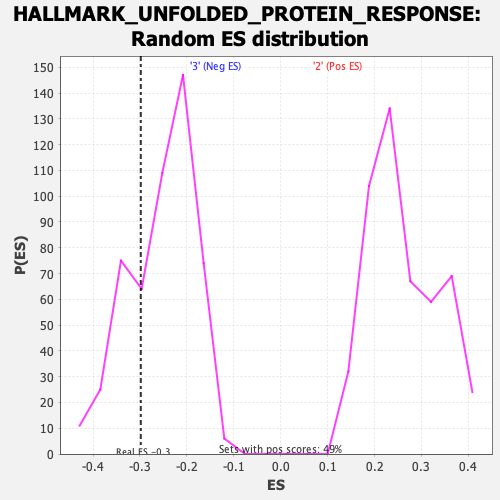
| Enrichment Score (ES) | -0.29813436 |
| Normalized Enrichment Score (NES) | -1.1691304 |
| Nominal p-value | 0.27201566 |
| FDR q-value | 0.73317313 |
| FWER p-Value | 0.956 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Psat1 | 64 | 0.924 | 0.0303 | No |
| 2 | Ddit4 | 418 | 0.628 | 0.0322 | No |
| 3 | Nop56 | 596 | 0.578 | 0.0429 | No |
| 4 | H2ax | 676 | 0.561 | 0.0589 | No |
| 5 | Mtrex | 797 | 0.535 | 0.0714 | No |
| 6 | Lsm4 | 962 | 0.498 | 0.0800 | No |
| 7 | Cebpg | 987 | 0.493 | 0.0967 | No |
| 8 | Srpr | 1201 | 0.457 | 0.1008 | No |
| 9 | Nfyb | 1779 | 0.394 | 0.0806 | No |
| 10 | Dkc1 | 1786 | 0.393 | 0.0948 | No |
| 11 | Cnot6 | 2066 | 0.361 | 0.0913 | No |
| 12 | Exosc10 | 2166 | 0.349 | 0.0982 | No |
| 13 | Nfya | 2673 | 0.306 | 0.0790 | No |
| 14 | Eif4a1 | 2808 | 0.294 | 0.0818 | No |
| 15 | Slc30a5 | 2933 | 0.284 | 0.0849 | No |
| 16 | Dnaja4 | 3044 | 0.274 | 0.0883 | No |
| 17 | Eif4ebp1 | 3094 | 0.271 | 0.0954 | No |
| 18 | Hspa9 | 3511 | 0.247 | 0.0795 | No |
| 19 | Slc7a5 | 3577 | 0.245 | 0.0846 | No |
| 20 | Eif4a2 | 3597 | 0.243 | 0.0924 | No |
| 21 | Ern1 | 4041 | 0.216 | 0.0737 | No |
| 22 | Eif4e | 4463 | 0.192 | 0.0554 | No |
| 23 | Cnot4 | 4478 | 0.191 | 0.0616 | No |
| 24 | Exosc2 | 4608 | 0.183 | 0.0606 | No |
| 25 | Gosr2 | 4611 | 0.183 | 0.0673 | No |
| 26 | Yif1a | 4621 | 0.182 | 0.0734 | No |
| 27 | Rps14 | 4756 | 0.173 | 0.0718 | No |
| 28 | Fus | 4933 | 0.164 | 0.0672 | No |
| 29 | Sec11a | 6697 | 0.074 | -0.0363 | No |
| 30 | Kif5b | 6732 | 0.072 | -0.0357 | No |
| 31 | Xbp1 | 6745 | 0.072 | -0.0337 | No |
| 32 | Wfs1 | 7284 | 0.048 | -0.0644 | No |
| 33 | Herpud1 | 7550 | 0.036 | -0.0790 | No |
| 34 | Aldh18a1 | 7778 | 0.025 | -0.0917 | No |
| 35 | Mthfd2 | 8342 | 0.001 | -0.1256 | No |
| 36 | Exosc4 | 8579 | -0.006 | -0.1396 | No |
| 37 | Xpot | 8812 | -0.016 | -0.1530 | No |
| 38 | Tubb2a | 9007 | -0.024 | -0.1638 | No |
| 39 | Dcp1a | 9035 | -0.025 | -0.1645 | No |
| 40 | Bag3 | 9053 | -0.025 | -0.1646 | No |
| 41 | Eif4a3 | 9054 | -0.025 | -0.1637 | No |
| 42 | Exosc9 | 9106 | -0.027 | -0.1658 | No |
| 43 | Npm1 | 9172 | -0.030 | -0.1686 | No |
| 44 | Parn | 9238 | -0.033 | -0.1712 | No |
| 45 | Dnajb9 | 9361 | -0.040 | -0.1771 | No |
| 46 | Eif2ak3 | 9848 | -0.062 | -0.2041 | No |
| 47 | Ssr1 | 9871 | -0.063 | -0.2031 | No |
| 48 | Cnot2 | 9913 | -0.065 | -0.2032 | No |
| 49 | Nhp2 | 10210 | -0.078 | -0.2181 | No |
| 50 | Exosc5 | 10234 | -0.079 | -0.2166 | No |
| 51 | Dcp2 | 10332 | -0.085 | -0.2193 | No |
| 52 | Shc1 | 10369 | -0.086 | -0.2183 | No |
| 53 | Pdia5 | 10516 | -0.093 | -0.2236 | No |
| 54 | Atp6v0d1 | 10583 | -0.096 | -0.2241 | No |
| 55 | Imp3 | 10834 | -0.107 | -0.2352 | No |
| 56 | Exoc2 | 11189 | -0.123 | -0.2520 | No |
| 57 | Nabp1 | 11245 | -0.127 | -0.2506 | No |
| 58 | Spcs1 | 11569 | -0.141 | -0.2649 | No |
| 59 | Rrp9 | 11713 | -0.148 | -0.2680 | No |
| 60 | Cks1b | 11732 | -0.150 | -0.2635 | No |
| 61 | Calr | 11906 | -0.157 | -0.2682 | No |
| 62 | Gemin4 | 11983 | -0.161 | -0.2668 | No |
| 63 | Tspyl2 | 12295 | -0.176 | -0.2790 | No |
| 64 | Vegfa | 12373 | -0.180 | -0.2770 | No |
| 65 | Banf1 | 12686 | -0.195 | -0.2886 | No |
| 66 | Arfgap1 | 12743 | -0.199 | -0.2847 | No |
| 67 | Asns | 12789 | -0.202 | -0.2799 | No |
| 68 | Paip1 | 12870 | -0.207 | -0.2771 | No |
| 69 | Pdia6 | 12966 | -0.213 | -0.2749 | No |
| 70 | Nolc1 | 13281 | -0.232 | -0.2853 | No |
| 71 | Cebpb | 13495 | -0.245 | -0.2891 | Yes |
| 72 | Tatdn2 | 13579 | -0.249 | -0.2849 | Yes |
| 73 | Eif2s1 | 13606 | -0.251 | -0.2772 | Yes |
| 74 | Khsrp | 13702 | -0.258 | -0.2734 | Yes |
| 75 | Ero1a | 13764 | -0.261 | -0.2674 | Yes |
| 76 | Dctn1 | 14126 | -0.289 | -0.2785 | Yes |
| 77 | Nop14 | 14210 | -0.296 | -0.2726 | Yes |
| 78 | Ywhaz | 14382 | -0.308 | -0.2715 | Yes |
| 79 | Zbtb17 | 14492 | -0.315 | -0.2664 | Yes |
| 80 | Eef2 | 14526 | -0.319 | -0.2566 | Yes |
| 81 | Edem1 | 14626 | -0.326 | -0.2506 | Yes |
| 82 | Dnajc3 | 14671 | -0.329 | -0.2411 | Yes |
| 83 | Atf6 | 14674 | -0.329 | -0.2290 | Yes |
| 84 | Hspa5 | 14817 | -0.345 | -0.2248 | Yes |
| 85 | Srprb | 14899 | -0.353 | -0.2167 | Yes |
| 86 | Sec31a | 14903 | -0.353 | -0.2038 | Yes |
| 87 | Eif4g1 | 14998 | -0.363 | -0.1960 | Yes |
| 88 | Wipi1 | 15145 | -0.376 | -0.1909 | Yes |
| 89 | Preb | 15188 | -0.380 | -0.1794 | Yes |
| 90 | Pop4 | 15242 | -0.389 | -0.1682 | Yes |
| 91 | Fkbp14 | 15333 | -0.400 | -0.1589 | Yes |
| 92 | Ddx10 | 15758 | -0.454 | -0.1677 | Yes |
| 93 | Hyou1 | 15772 | -0.456 | -0.1516 | Yes |
| 94 | Sdad1 | 15800 | -0.461 | -0.1362 | Yes |
| 95 | Exosc1 | 15842 | -0.469 | -0.1213 | Yes |
| 96 | Edc4 | 15912 | -0.482 | -0.1077 | Yes |
| 97 | Chac1 | 16097 | -0.516 | -0.0997 | Yes |
| 98 | Hsp90b1 | 16159 | -0.530 | -0.0838 | Yes |
| 99 | Atf4 | 16217 | -0.549 | -0.0669 | Yes |
| 100 | Cxxc1 | 16276 | -0.568 | -0.0494 | Yes |
| 101 | Slc1a4 | 16305 | -0.579 | -0.0297 | Yes |
| 102 | Serp1 | 16483 | -0.664 | -0.0158 | Yes |
| 103 | Lsm1 | 16609 | -0.779 | 0.0054 | Yes |