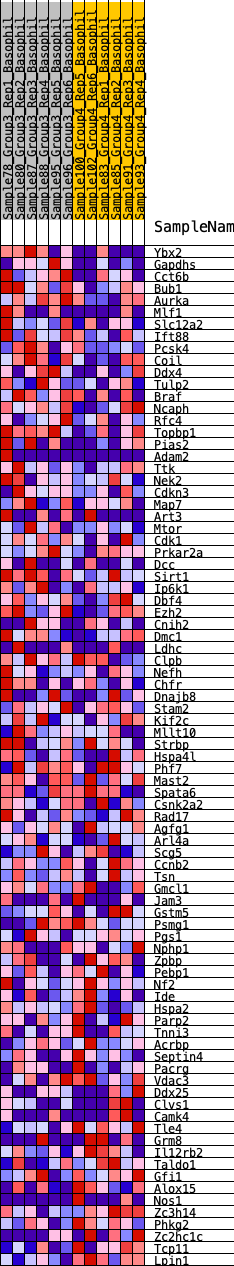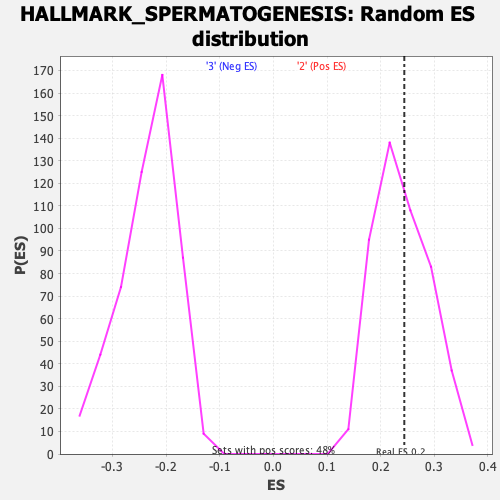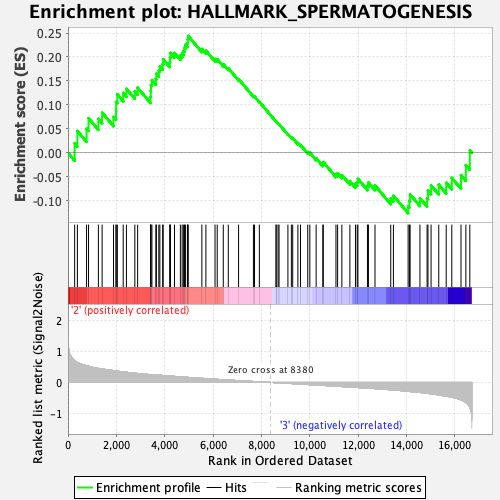
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group3_versus_Group4.Basophil_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_SPERMATOGENESIS |
| Enrichment Score (ES) | 0.2440879 |
| Normalized Enrichment Score (NES) | 1.0180923 |
| Nominal p-value | 0.42857143 |
| FDR q-value | 0.98645777 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ybx2 | 275 | 0.707 | 0.0196 | Yes |
| 2 | Gapdhs | 388 | 0.638 | 0.0454 | Yes |
| 3 | Cct6b | 767 | 0.542 | 0.0503 | Yes |
| 4 | Bub1 | 849 | 0.524 | 0.0722 | Yes |
| 5 | Aurka | 1256 | 0.452 | 0.0709 | Yes |
| 6 | Mlf1 | 1411 | 0.433 | 0.0837 | Yes |
| 7 | Slc12a2 | 1883 | 0.382 | 0.0749 | Yes |
| 8 | Ift88 | 1985 | 0.371 | 0.0877 | Yes |
| 9 | Pcsk4 | 1991 | 0.370 | 0.1064 | Yes |
| 10 | Coil | 2041 | 0.364 | 0.1220 | Yes |
| 11 | Ddx4 | 2283 | 0.339 | 0.1248 | Yes |
| 12 | Tulp2 | 2416 | 0.328 | 0.1336 | Yes |
| 13 | Braf | 2764 | 0.298 | 0.1280 | Yes |
| 14 | Ncaph | 2880 | 0.289 | 0.1358 | Yes |
| 15 | Rfc4 | 3412 | 0.252 | 0.1167 | Yes |
| 16 | Topbp1 | 3422 | 0.251 | 0.1290 | Yes |
| 17 | Pias2 | 3430 | 0.251 | 0.1414 | Yes |
| 18 | Adam2 | 3470 | 0.248 | 0.1517 | Yes |
| 19 | Ttk | 3634 | 0.241 | 0.1542 | Yes |
| 20 | Nek2 | 3654 | 0.239 | 0.1653 | Yes |
| 21 | Cdkn3 | 3756 | 0.235 | 0.1712 | Yes |
| 22 | Map7 | 3799 | 0.232 | 0.1805 | Yes |
| 23 | Art3 | 3916 | 0.225 | 0.1850 | Yes |
| 24 | Mtor | 3935 | 0.224 | 0.1954 | Yes |
| 25 | Cdk1 | 4211 | 0.204 | 0.1892 | Yes |
| 26 | Prkar2a | 4219 | 0.203 | 0.1992 | Yes |
| 27 | Dcc | 4236 | 0.203 | 0.2086 | Yes |
| 28 | Sirt1 | 4404 | 0.194 | 0.2084 | Yes |
| 29 | Ip6k1 | 4647 | 0.180 | 0.2031 | Yes |
| 30 | Dbf4 | 4720 | 0.175 | 0.2077 | Yes |
| 31 | Ezh2 | 4782 | 0.172 | 0.2128 | Yes |
| 32 | Cnih2 | 4819 | 0.171 | 0.2193 | Yes |
| 33 | Dmc1 | 4855 | 0.168 | 0.2258 | Yes |
| 34 | Ldhc | 4938 | 0.164 | 0.2292 | Yes |
| 35 | Clpb | 4954 | 0.163 | 0.2366 | Yes |
| 36 | Nefh | 4969 | 0.162 | 0.2441 | Yes |
| 37 | Chfr | 5536 | 0.133 | 0.2168 | No |
| 38 | Dnajb8 | 5702 | 0.124 | 0.2132 | No |
| 39 | Stam2 | 6080 | 0.105 | 0.1959 | No |
| 40 | Kif2c | 6171 | 0.100 | 0.1955 | No |
| 41 | Mllt10 | 6421 | 0.088 | 0.1850 | No |
| 42 | Strbp | 6628 | 0.077 | 0.1766 | No |
| 43 | Hspa4l | 7054 | 0.058 | 0.1539 | No |
| 44 | Phf7 | 7668 | 0.031 | 0.1186 | No |
| 45 | Mast2 | 7714 | 0.029 | 0.1174 | No |
| 46 | Spata6 | 7913 | 0.020 | 0.1065 | No |
| 47 | Csnk2a2 | 8604 | -0.007 | 0.0653 | No |
| 48 | Rad17 | 8608 | -0.007 | 0.0655 | No |
| 49 | Agfg1 | 8686 | -0.010 | 0.0614 | No |
| 50 | Arl4a | 8727 | -0.012 | 0.0596 | No |
| 51 | Scg5 | 9094 | -0.027 | 0.0390 | No |
| 52 | Ccnb2 | 9237 | -0.033 | 0.0321 | No |
| 53 | Tsn | 9292 | -0.036 | 0.0307 | No |
| 54 | Gmcl1 | 9507 | -0.046 | 0.0202 | No |
| 55 | Jam3 | 9615 | -0.051 | 0.0164 | No |
| 56 | Gstm5 | 9915 | -0.065 | 0.0017 | No |
| 57 | Psmg1 | 10000 | -0.069 | 0.0002 | No |
| 58 | Pgs1 | 10261 | -0.080 | -0.0114 | No |
| 59 | Nphp1 | 10538 | -0.094 | -0.0232 | No |
| 60 | Zpbp | 10557 | -0.094 | -0.0195 | No |
| 61 | Pebp1 | 11081 | -0.118 | -0.0449 | No |
| 62 | Nf2 | 11149 | -0.122 | -0.0427 | No |
| 63 | Ide | 11325 | -0.130 | -0.0466 | No |
| 64 | Hspa2 | 11657 | -0.146 | -0.0590 | No |
| 65 | Parp2 | 11887 | -0.156 | -0.0649 | No |
| 66 | Tnni3 | 11959 | -0.160 | -0.0610 | No |
| 67 | Acrbp | 11990 | -0.161 | -0.0546 | No |
| 68 | Septin4 | 12384 | -0.181 | -0.0690 | No |
| 69 | Pacrg | 12424 | -0.183 | -0.0620 | No |
| 70 | Vdac3 | 12695 | -0.196 | -0.0682 | No |
| 71 | Ddx25 | 13346 | -0.236 | -0.0953 | No |
| 72 | Clvs1 | 13457 | -0.242 | -0.0896 | No |
| 73 | Camk4 | 14065 | -0.283 | -0.1116 | No |
| 74 | Tle4 | 14118 | -0.288 | -0.1001 | No |
| 75 | Grm8 | 14147 | -0.291 | -0.0869 | No |
| 76 | Il12rb2 | 14554 | -0.321 | -0.0949 | No |
| 77 | Taldo1 | 14849 | -0.348 | -0.0949 | No |
| 78 | Gfi1 | 14886 | -0.351 | -0.0791 | No |
| 79 | Alox15 | 15014 | -0.365 | -0.0681 | No |
| 80 | Nos1 | 15332 | -0.400 | -0.0667 | No |
| 81 | Zc3h14 | 15640 | -0.440 | -0.0627 | No |
| 82 | Phkg2 | 15872 | -0.474 | -0.0524 | No |
| 83 | Zc2hc1c | 16252 | -0.559 | -0.0467 | No |
| 84 | Tcp11 | 16453 | -0.645 | -0.0258 | No |
| 85 | Lpin1 | 16617 | -0.794 | 0.0049 | No |