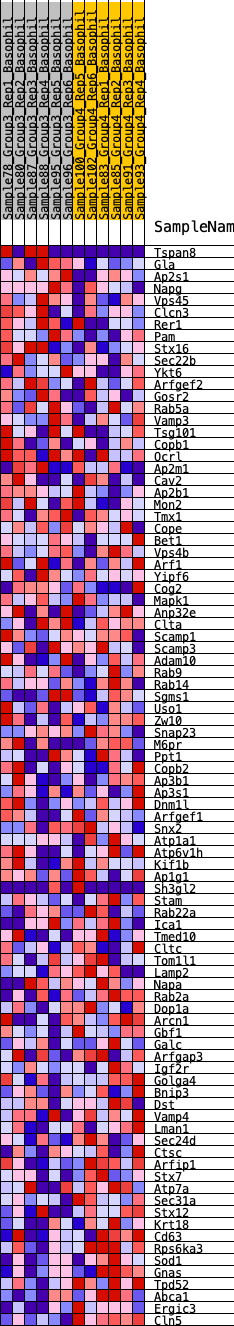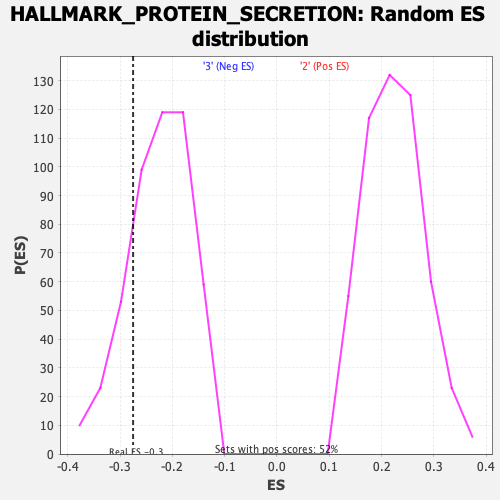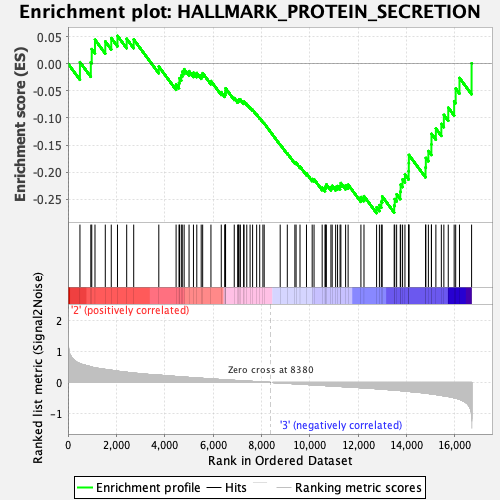
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group3_versus_Group4.Basophil_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_PROTEIN_SECRETION |
| Enrichment Score (ES) | -0.27523234 |
| Normalized Enrichment Score (NES) | -1.2164023 |
| Nominal p-value | 0.20124482 |
| FDR q-value | 0.65872717 |
| FWER p-Value | 0.923 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Tspan8 | 494 | 0.608 | 0.0028 | No |
| 2 | Gla | 944 | 0.501 | 0.0026 | No |
| 3 | Ap2s1 | 982 | 0.495 | 0.0269 | No |
| 4 | Napg | 1116 | 0.471 | 0.0441 | No |
| 5 | Vps45 | 1542 | 0.419 | 0.0409 | No |
| 6 | Clcn3 | 1789 | 0.392 | 0.0471 | No |
| 7 | Rer1 | 2046 | 0.363 | 0.0511 | No |
| 8 | Pam | 2427 | 0.327 | 0.0458 | No |
| 9 | Stx16 | 2717 | 0.302 | 0.0445 | No |
| 10 | Sec22b | 3755 | 0.235 | -0.0053 | No |
| 11 | Ykt6 | 4470 | 0.191 | -0.0381 | No |
| 12 | Arfgef2 | 4591 | 0.184 | -0.0355 | No |
| 13 | Gosr2 | 4611 | 0.183 | -0.0268 | No |
| 14 | Rab5a | 4685 | 0.177 | -0.0217 | No |
| 15 | Vamp3 | 4725 | 0.175 | -0.0147 | No |
| 16 | Tsg101 | 4803 | 0.171 | -0.0102 | No |
| 17 | Copb1 | 5006 | 0.161 | -0.0138 | No |
| 18 | Ocrl | 5188 | 0.149 | -0.0167 | No |
| 19 | Ap2m1 | 5326 | 0.144 | -0.0172 | No |
| 20 | Cav2 | 5508 | 0.134 | -0.0209 | No |
| 21 | Ap2b1 | 5569 | 0.131 | -0.0175 | No |
| 22 | Mon2 | 5911 | 0.113 | -0.0320 | No |
| 23 | Tmx1 | 6338 | 0.092 | -0.0527 | No |
| 24 | Cope | 6481 | 0.085 | -0.0567 | No |
| 25 | Bet1 | 6505 | 0.083 | -0.0537 | No |
| 26 | Vps4b | 6513 | 0.083 | -0.0497 | No |
| 27 | Arf1 | 6516 | 0.083 | -0.0454 | No |
| 28 | Yipf6 | 6875 | 0.066 | -0.0634 | No |
| 29 | Cog2 | 7017 | 0.060 | -0.0687 | No |
| 30 | Mapk1 | 7048 | 0.058 | -0.0674 | No |
| 31 | Anp32e | 7073 | 0.057 | -0.0658 | No |
| 32 | Clta | 7129 | 0.055 | -0.0661 | No |
| 33 | Scamp1 | 7263 | 0.049 | -0.0715 | No |
| 34 | Scamp3 | 7279 | 0.048 | -0.0698 | No |
| 35 | Adam10 | 7393 | 0.043 | -0.0743 | No |
| 36 | Rab9 | 7530 | 0.037 | -0.0805 | No |
| 37 | Rab14 | 7634 | 0.032 | -0.0850 | No |
| 38 | Sgms1 | 7801 | 0.024 | -0.0937 | No |
| 39 | Uso1 | 7928 | 0.019 | -0.1002 | No |
| 40 | Zw10 | 8060 | 0.014 | -0.1074 | No |
| 41 | Snap23 | 8113 | 0.011 | -0.1099 | No |
| 42 | M6pr | 8775 | -0.015 | -0.1489 | No |
| 43 | Ppt1 | 9069 | -0.026 | -0.1652 | No |
| 44 | Copb2 | 9382 | -0.041 | -0.1818 | No |
| 45 | Ap3b1 | 9432 | -0.043 | -0.1824 | No |
| 46 | Ap3s1 | 9594 | -0.050 | -0.1894 | No |
| 47 | Dnm1l | 9862 | -0.062 | -0.2022 | No |
| 48 | Arfgef1 | 10098 | -0.073 | -0.2124 | No |
| 49 | Snx2 | 10175 | -0.077 | -0.2128 | No |
| 50 | Atp1a1 | 10511 | -0.092 | -0.2281 | No |
| 51 | Atp6v1h | 10628 | -0.098 | -0.2298 | No |
| 52 | Kif1b | 10652 | -0.099 | -0.2259 | No |
| 53 | Ap1g1 | 10684 | -0.101 | -0.2224 | No |
| 54 | Sh3gl2 | 10873 | -0.109 | -0.2278 | No |
| 55 | Stam | 10924 | -0.112 | -0.2248 | No |
| 56 | Rab22a | 11068 | -0.118 | -0.2271 | No |
| 57 | Ica1 | 11147 | -0.122 | -0.2253 | No |
| 58 | Tmed10 | 11251 | -0.127 | -0.2247 | No |
| 59 | Cltc | 11284 | -0.128 | -0.2198 | No |
| 60 | Tom1l1 | 11480 | -0.138 | -0.2242 | No |
| 61 | Lamp2 | 11584 | -0.142 | -0.2227 | No |
| 62 | Napa | 12113 | -0.167 | -0.2456 | No |
| 63 | Rab2a | 12240 | -0.173 | -0.2439 | No |
| 64 | Dop1a | 12761 | -0.200 | -0.2645 | Yes |
| 65 | Arcn1 | 12878 | -0.207 | -0.2604 | Yes |
| 66 | Gbf1 | 12958 | -0.212 | -0.2538 | Yes |
| 67 | Galc | 12993 | -0.214 | -0.2444 | Yes |
| 68 | Arfgap3 | 13489 | -0.244 | -0.2611 | Yes |
| 69 | Igf2r | 13512 | -0.246 | -0.2493 | Yes |
| 70 | Golga4 | 13590 | -0.250 | -0.2405 | Yes |
| 71 | Bnip3 | 13740 | -0.260 | -0.2356 | Yes |
| 72 | Dst | 13757 | -0.261 | -0.2226 | Yes |
| 73 | Vamp4 | 13836 | -0.266 | -0.2131 | Yes |
| 74 | Lman1 | 13934 | -0.273 | -0.2043 | Yes |
| 75 | Sec24d | 14086 | -0.285 | -0.1981 | Yes |
| 76 | Ctsc | 14093 | -0.286 | -0.1832 | Yes |
| 77 | Arfip1 | 14097 | -0.286 | -0.1680 | Yes |
| 78 | Stx7 | 14785 | -0.341 | -0.1911 | Yes |
| 79 | Atp7a | 14798 | -0.342 | -0.1735 | Yes |
| 80 | Sec31a | 14903 | -0.353 | -0.1608 | Yes |
| 81 | Stx12 | 15027 | -0.366 | -0.1486 | Yes |
| 82 | Krt18 | 15032 | -0.367 | -0.1292 | Yes |
| 83 | Cd63 | 15213 | -0.383 | -0.1196 | Yes |
| 84 | Rps6ka3 | 15439 | -0.414 | -0.1109 | Yes |
| 85 | Sod1 | 15543 | -0.427 | -0.0943 | Yes |
| 86 | Gnas | 15725 | -0.452 | -0.0809 | Yes |
| 87 | Tpd52 | 15969 | -0.492 | -0.0692 | Yes |
| 88 | Abca1 | 16032 | -0.507 | -0.0458 | Yes |
| 89 | Ergic3 | 16189 | -0.539 | -0.0263 | Yes |
| 90 | Cln5 | 16690 | -1.064 | 0.0005 | Yes |