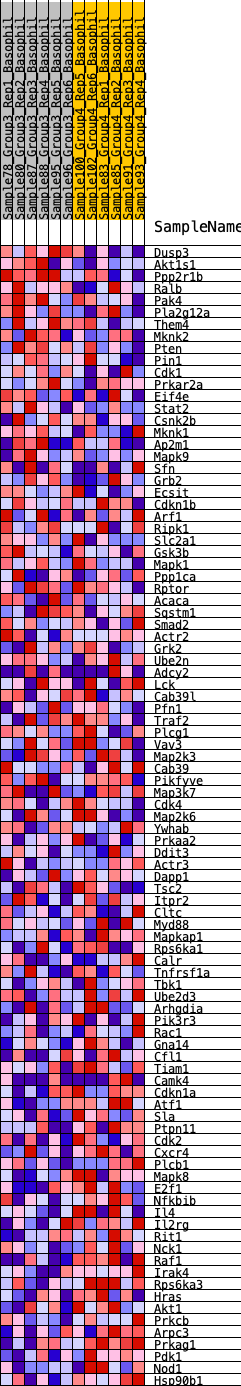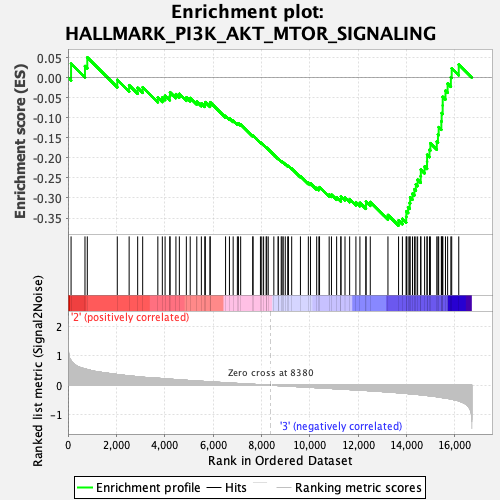
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group3_versus_Group4.Basophil_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_PI3K_AKT_MTOR_SIGNALING |
| Enrichment Score (ES) | -0.36879987 |
| Normalized Enrichment Score (NES) | -1.6815156 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.1916635 |
| FWER p-Value | 0.119 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Dusp3 | 127 | 0.836 | 0.0350 | No |
| 2 | Akt1s1 | 702 | 0.555 | 0.0287 | No |
| 3 | Ppp2r1b | 798 | 0.535 | 0.0503 | No |
| 4 | Ralb | 2038 | 0.364 | -0.0058 | No |
| 5 | Pak4 | 2528 | 0.317 | -0.0191 | No |
| 6 | Pla2g12a | 2881 | 0.289 | -0.0255 | No |
| 7 | Them4 | 3088 | 0.271 | -0.0241 | No |
| 8 | Mknk2 | 3715 | 0.237 | -0.0497 | No |
| 9 | Pten | 3901 | 0.226 | -0.0494 | No |
| 10 | Pin1 | 4015 | 0.218 | -0.0451 | No |
| 11 | Cdk1 | 4211 | 0.204 | -0.0464 | No |
| 12 | Prkar2a | 4219 | 0.203 | -0.0365 | No |
| 13 | Eif4e | 4463 | 0.192 | -0.0413 | No |
| 14 | Stat2 | 4605 | 0.183 | -0.0405 | No |
| 15 | Csnk2b | 4894 | 0.166 | -0.0494 | No |
| 16 | Mknk1 | 5054 | 0.158 | -0.0509 | No |
| 17 | Ap2m1 | 5326 | 0.144 | -0.0599 | No |
| 18 | Mapk9 | 5515 | 0.134 | -0.0644 | No |
| 19 | Sfn | 5658 | 0.126 | -0.0665 | No |
| 20 | Grb2 | 5678 | 0.125 | -0.0612 | No |
| 21 | Ecsit | 5874 | 0.115 | -0.0671 | No |
| 22 | Cdkn1b | 5878 | 0.115 | -0.0614 | No |
| 23 | Arf1 | 6516 | 0.083 | -0.0956 | No |
| 24 | Ripk1 | 6676 | 0.075 | -0.1014 | No |
| 25 | Slc2a1 | 6830 | 0.068 | -0.1071 | No |
| 26 | Gsk3b | 7004 | 0.060 | -0.1144 | No |
| 27 | Mapk1 | 7048 | 0.058 | -0.1141 | No |
| 28 | Ppp1ca | 7140 | 0.055 | -0.1168 | No |
| 29 | Rptor | 7639 | 0.032 | -0.1451 | No |
| 30 | Acaca | 7653 | 0.032 | -0.1443 | No |
| 31 | Sqstm1 | 7958 | 0.018 | -0.1617 | No |
| 32 | Smad2 | 8013 | 0.015 | -0.1642 | No |
| 33 | Actr2 | 8089 | 0.012 | -0.1680 | No |
| 34 | Grk2 | 8194 | 0.008 | -0.1739 | No |
| 35 | Ube2n | 8218 | 0.007 | -0.1749 | No |
| 36 | Adcy2 | 8280 | 0.004 | -0.1784 | No |
| 37 | Lck | 8512 | -0.002 | -0.1922 | No |
| 38 | Cab39l | 8692 | -0.011 | -0.2024 | No |
| 39 | Pfn1 | 8693 | -0.011 | -0.2019 | No |
| 40 | Traf2 | 8817 | -0.016 | -0.2085 | No |
| 41 | Plcg1 | 8852 | -0.018 | -0.2096 | No |
| 42 | Vav3 | 8900 | -0.019 | -0.2115 | No |
| 43 | Map2k3 | 8985 | -0.023 | -0.2154 | No |
| 44 | Cab39 | 9087 | -0.027 | -0.2201 | No |
| 45 | Pikfyve | 9116 | -0.028 | -0.2204 | No |
| 46 | Map3k7 | 9250 | -0.034 | -0.2266 | No |
| 47 | Cdk4 | 9612 | -0.051 | -0.2458 | No |
| 48 | Map2k6 | 9939 | -0.066 | -0.2620 | No |
| 49 | Ywhab | 10027 | -0.070 | -0.2637 | No |
| 50 | Prkaa2 | 10284 | -0.082 | -0.2750 | No |
| 51 | Ddit3 | 10376 | -0.086 | -0.2760 | No |
| 52 | Actr3 | 10402 | -0.087 | -0.2731 | No |
| 53 | Dapp1 | 10800 | -0.106 | -0.2916 | No |
| 54 | Tsc2 | 10895 | -0.110 | -0.2916 | No |
| 55 | Itpr2 | 11109 | -0.120 | -0.2984 | No |
| 56 | Cltc | 11284 | -0.128 | -0.3023 | No |
| 57 | Myd88 | 11287 | -0.129 | -0.2959 | No |
| 58 | Mapkap1 | 11455 | -0.137 | -0.2989 | No |
| 59 | Rps6ka1 | 11647 | -0.145 | -0.3030 | No |
| 60 | Calr | 11906 | -0.157 | -0.3106 | No |
| 61 | Tnfrsf1a | 12072 | -0.165 | -0.3121 | No |
| 62 | Tbk1 | 12320 | -0.177 | -0.3179 | No |
| 63 | Ube2d3 | 12323 | -0.177 | -0.3090 | No |
| 64 | Arhgdia | 12500 | -0.188 | -0.3101 | No |
| 65 | Pik3r3 | 13231 | -0.228 | -0.3424 | No |
| 66 | Rac1 | 13670 | -0.255 | -0.3558 | Yes |
| 67 | Gna14 | 13829 | -0.265 | -0.3518 | Yes |
| 68 | Cfl1 | 13985 | -0.277 | -0.3470 | Yes |
| 69 | Tiam1 | 13992 | -0.278 | -0.3332 | Yes |
| 70 | Camk4 | 14065 | -0.283 | -0.3231 | Yes |
| 71 | Cdkn1a | 14130 | -0.289 | -0.3122 | Yes |
| 72 | Atf1 | 14149 | -0.291 | -0.2985 | Yes |
| 73 | Sla | 14246 | -0.299 | -0.2890 | Yes |
| 74 | Ptpn11 | 14326 | -0.305 | -0.2783 | Yes |
| 75 | Cdk2 | 14385 | -0.308 | -0.2661 | Yes |
| 76 | Cxcr4 | 14458 | -0.313 | -0.2545 | Yes |
| 77 | Plcb1 | 14585 | -0.323 | -0.2456 | Yes |
| 78 | Mapk8 | 14592 | -0.324 | -0.2294 | Yes |
| 79 | E2f1 | 14747 | -0.336 | -0.2216 | Yes |
| 80 | Nfkbib | 14848 | -0.348 | -0.2099 | Yes |
| 81 | Il4 | 14851 | -0.348 | -0.1923 | Yes |
| 82 | Il2rg | 14949 | -0.359 | -0.1798 | Yes |
| 83 | Rit1 | 14986 | -0.362 | -0.1635 | Yes |
| 84 | Nck1 | 15251 | -0.390 | -0.1595 | Yes |
| 85 | Raf1 | 15299 | -0.397 | -0.1421 | Yes |
| 86 | Irak4 | 15329 | -0.400 | -0.1235 | Yes |
| 87 | Rps6ka3 | 15439 | -0.414 | -0.1090 | Yes |
| 88 | Hras | 15452 | -0.416 | -0.0885 | Yes |
| 89 | Akt1 | 15494 | -0.422 | -0.0694 | Yes |
| 90 | Prkcb | 15496 | -0.422 | -0.0480 | Yes |
| 91 | Arpc3 | 15610 | -0.437 | -0.0325 | Yes |
| 92 | Prkag1 | 15699 | -0.449 | -0.0149 | Yes |
| 93 | Pdk1 | 15833 | -0.468 | 0.0009 | Yes |
| 94 | Nod1 | 15868 | -0.473 | 0.0230 | Yes |
| 95 | Hsp90b1 | 16159 | -0.530 | 0.0325 | Yes |