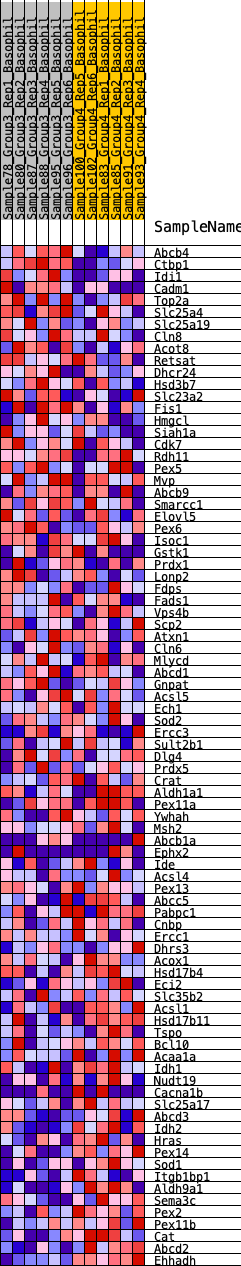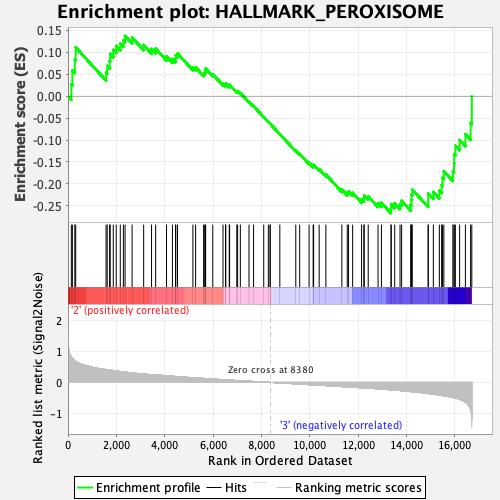
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group3_versus_Group4.Basophil_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_PEROXISOME |
| Enrichment Score (ES) | -0.2666459 |
| Normalized Enrichment Score (NES) | -1.0602523 |
| Nominal p-value | 0.36619717 |
| FDR q-value | 0.79265 |
| FWER p-Value | 0.99 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Abcb4 | 138 | 0.816 | 0.0274 | No |
| 2 | Ctbp1 | 181 | 0.772 | 0.0587 | No |
| 3 | Idi1 | 282 | 0.704 | 0.0835 | No |
| 4 | Cadm1 | 319 | 0.678 | 0.1111 | No |
| 5 | Top2a | 1574 | 0.416 | 0.0538 | No |
| 6 | Slc25a4 | 1621 | 0.411 | 0.0690 | No |
| 7 | Slc25a19 | 1721 | 0.398 | 0.0805 | No |
| 8 | Cln8 | 1745 | 0.395 | 0.0965 | No |
| 9 | Acot8 | 1872 | 0.383 | 0.1057 | No |
| 10 | Retsat | 1995 | 0.370 | 0.1145 | No |
| 11 | Dhcr24 | 2162 | 0.350 | 0.1199 | No |
| 12 | Hsd3b7 | 2290 | 0.338 | 0.1270 | No |
| 13 | Slc23a2 | 2360 | 0.333 | 0.1375 | No |
| 14 | Fis1 | 2650 | 0.308 | 0.1335 | No |
| 15 | Hmgcl | 3129 | 0.269 | 0.1165 | No |
| 16 | Siah1a | 3455 | 0.249 | 0.1079 | No |
| 17 | Cdk7 | 3625 | 0.241 | 0.1083 | No |
| 18 | Rdh11 | 4074 | 0.213 | 0.0907 | No |
| 19 | Pex5 | 4319 | 0.200 | 0.0847 | No |
| 20 | Mvp | 4447 | 0.193 | 0.0855 | No |
| 21 | Abcb9 | 4458 | 0.192 | 0.0933 | No |
| 22 | Smarcc1 | 4529 | 0.187 | 0.0973 | No |
| 23 | Elovl5 | 5165 | 0.151 | 0.0657 | No |
| 24 | Pex6 | 5277 | 0.146 | 0.0654 | No |
| 25 | Isoc1 | 5608 | 0.129 | 0.0512 | No |
| 26 | Gstk1 | 5653 | 0.127 | 0.0541 | No |
| 27 | Prdx1 | 5679 | 0.125 | 0.0581 | No |
| 28 | Lonp2 | 5692 | 0.125 | 0.0628 | No |
| 29 | Fdps | 5987 | 0.109 | 0.0499 | No |
| 30 | Fads1 | 6410 | 0.088 | 0.0284 | No |
| 31 | Vps4b | 6513 | 0.083 | 0.0259 | No |
| 32 | Scp2 | 6526 | 0.082 | 0.0287 | No |
| 33 | Atxn1 | 6668 | 0.075 | 0.0235 | No |
| 34 | Cln6 | 6683 | 0.074 | 0.0259 | No |
| 35 | Mlycd | 6981 | 0.061 | 0.0107 | No |
| 36 | Abcd1 | 7014 | 0.060 | 0.0114 | No |
| 37 | Gnpat | 7127 | 0.055 | 0.0071 | No |
| 38 | Acsl5 | 7485 | 0.039 | -0.0127 | No |
| 39 | Ech1 | 7674 | 0.031 | -0.0227 | No |
| 40 | Sod2 | 8094 | 0.012 | -0.0474 | No |
| 41 | Ercc3 | 8285 | 0.004 | -0.0586 | No |
| 42 | Sult2b1 | 8358 | 0.001 | -0.0629 | No |
| 43 | Dlg4 | 8361 | 0.001 | -0.0630 | No |
| 44 | Prdx5 | 8760 | -0.014 | -0.0864 | No |
| 45 | Crat | 9420 | -0.043 | -0.1242 | No |
| 46 | Aldh1a1 | 9581 | -0.049 | -0.1316 | No |
| 47 | Pex11a | 9969 | -0.068 | -0.1520 | No |
| 48 | Ywhah | 10138 | -0.075 | -0.1588 | No |
| 49 | Msh2 | 10158 | -0.076 | -0.1566 | No |
| 50 | Abcb1a | 10386 | -0.087 | -0.1664 | No |
| 51 | Ephx2 | 10664 | -0.100 | -0.1787 | No |
| 52 | Ide | 11325 | -0.130 | -0.2127 | No |
| 53 | Acsl4 | 11548 | -0.140 | -0.2200 | No |
| 54 | Pex13 | 11601 | -0.143 | -0.2168 | No |
| 55 | Abcc5 | 11771 | -0.151 | -0.2204 | No |
| 56 | Pabpc1 | 12140 | -0.168 | -0.2351 | No |
| 57 | Cnbp | 12235 | -0.173 | -0.2332 | No |
| 58 | Ercc1 | 12249 | -0.174 | -0.2264 | No |
| 59 | Dhrs3 | 12415 | -0.182 | -0.2284 | No |
| 60 | Acox1 | 12821 | -0.204 | -0.2438 | No |
| 61 | Hsd17b4 | 12961 | -0.212 | -0.2429 | No |
| 62 | Eci2 | 13357 | -0.236 | -0.2563 | Yes |
| 63 | Slc35b2 | 13367 | -0.237 | -0.2464 | Yes |
| 64 | Acsl1 | 13511 | -0.246 | -0.2443 | Yes |
| 65 | Hsd17b11 | 13731 | -0.259 | -0.2461 | Yes |
| 66 | Tspo | 13796 | -0.263 | -0.2384 | Yes |
| 67 | Bcl10 | 14171 | -0.293 | -0.2481 | Yes |
| 68 | Acaa1a | 14200 | -0.295 | -0.2369 | Yes |
| 69 | Idh1 | 14205 | -0.295 | -0.2242 | Yes |
| 70 | Nudt19 | 14237 | -0.298 | -0.2130 | Yes |
| 71 | Cacna1b | 14895 | -0.352 | -0.2371 | Yes |
| 72 | Slc25a17 | 14897 | -0.353 | -0.2217 | Yes |
| 73 | Abcd3 | 15108 | -0.373 | -0.2180 | Yes |
| 74 | Idh2 | 15356 | -0.403 | -0.2153 | Yes |
| 75 | Hras | 15452 | -0.416 | -0.2028 | Yes |
| 76 | Pex14 | 15486 | -0.421 | -0.1863 | Yes |
| 77 | Sod1 | 15543 | -0.427 | -0.1710 | Yes |
| 78 | Itgb1bp1 | 15915 | -0.482 | -0.1722 | Yes |
| 79 | Aldh9a1 | 15964 | -0.492 | -0.1535 | Yes |
| 80 | Sema3c | 15971 | -0.493 | -0.1323 | Yes |
| 81 | Pex2 | 16018 | -0.504 | -0.1130 | Yes |
| 82 | Pex11b | 16195 | -0.540 | -0.0999 | Yes |
| 83 | Cat | 16434 | -0.632 | -0.0866 | Yes |
| 84 | Abcd2 | 16649 | -0.868 | -0.0614 | Yes |
| 85 | Ehhadh | 16699 | -1.470 | -0.0000 | Yes |